Chapter 13. Adventures In Covariance
- this chapter will show how to specify varying slopes in
combination with varying intercepts
- enables pooling to improve estimates of how different units respond to or are influenced by predictor variables
- also improves the estimates of intercepts by “borrowing information across parameter types”
- “varying slope models are massive interaction machines”
13.1 Varying slopes by construction
- pool information across intercepts and slopes by modeling the joint
population of intercepts and slopes
- modeling their covariance
- assigning a 2D Gaussian distribution to both the intercepts (first dimension) and the slopes (second dimension)
- the variance-covariance matrix for a fit model describes how each
parameters posterior probability is associated with one another
- varying intercepts have variation, varying slopes have variation, and intercepts and slopes covary
- use example of visiting coffee shops:
- visit different cafes, order a coffee, and record the wait time
- previously, used varying intercepts, one for each cafe
- also record the time of day
- the average wait time is longer in the mornings than afternoons because they are busier in the mornings
- different cafes vary in their average wait times and their
differences between morning and afternoon
- the differences in wait time by time of day are the slopes
- cafes covary in their intercepts and slopes
- because the popular cafes have much longer wait times in the morning leading to large differences between morning and afternoon
- visit different cafes, order a coffee, and record the wait time
$$ \mu_i = \alpha_{\text{cafe}[i]} + \beta_{\text{cafe}[i]} A_i $$
13.1.1 Simulate the population
- define the population of cafes
- define the average wait time in the morning and afternoon
- define the correlation between them
a <- 3.5 # average morning wait time
b <- -1 # average difference afternoon wait time
sigma_a <- 1 # standard deviation in intercepts
sigma_b <- 0.5 # standard deviation in slopes
rho <- -0.7 # correlation between intercepts and slopes
- use these values to simulate a sample of cafes
- define the multivariate Gaussian with a vector of means and a 2x2 matrix of variances and covariances
Mu <- c(a, b) # vector of two means
- the matrix of variances and covariances is arranged as follows
$$ \begin{pmatrix} \sigma_\alpha^2 & \sigma_\alpha \sigma_\beta \rho $$ $$ \sigma_\alpha \sigma_\beta \rho & \sigma_\beta^2 \end{pmatrix} $$
- can construct the matrix explicitly
cov_ab <- sigma_a * sigma_b * rho
Sigma <- matrix(c(sigma_a^2, cov_ab, cov_ab, sigma_b^2), nrow = 2)
Sigma
#> [,1] [,2]
#> [1,] 1.00 -0.35
#> [2,] -0.35 0.25
- another way to build the variance-covariance matrix using matrix
multiplication
- this is likely a better approach with larger models
sigmas <- c(sigma_a, sigma_b)
Rho <- matrix(c(1, rho, rho, 1), nrow = 2)
Sigma <- diag(sigmas) %*% Rho %*% diag(sigmas)
Sigma
#> [,1] [,2]
#> [1,] 1.00 -0.35
#> [2,] -0.35 0.25
- now can simulate 20 cafes, each with their own intercept and slope
N_cafes <- 20
- simulate from a multivariate Gaussian using
mvnorm()from the ‘MASS’ package- returns a matrix of $\text{cafe} \times (\text{intercept}, \text{slope})$
library(MASS)
set.seed(5)
vary_effects <- mvrnorm(n = N_cafes, mu = Mu, Sigma = Sigma)
head(vary_effects)
#> [,1] [,2]
#> [1,] 4.223962 -1.6093565
#> [2,] 2.010498 -0.7517704
#> [3,] 4.565811 -1.9482646
#> [4,] 3.343635 -1.1926539
#> [5,] 1.700971 -0.5855618
#> [6,] 4.134373 -1.1444539
# Split into separate vectors for ease of use later.
a_cafe <- vary_effects[, 1]
b_cafe <- vary_effects[, 2]
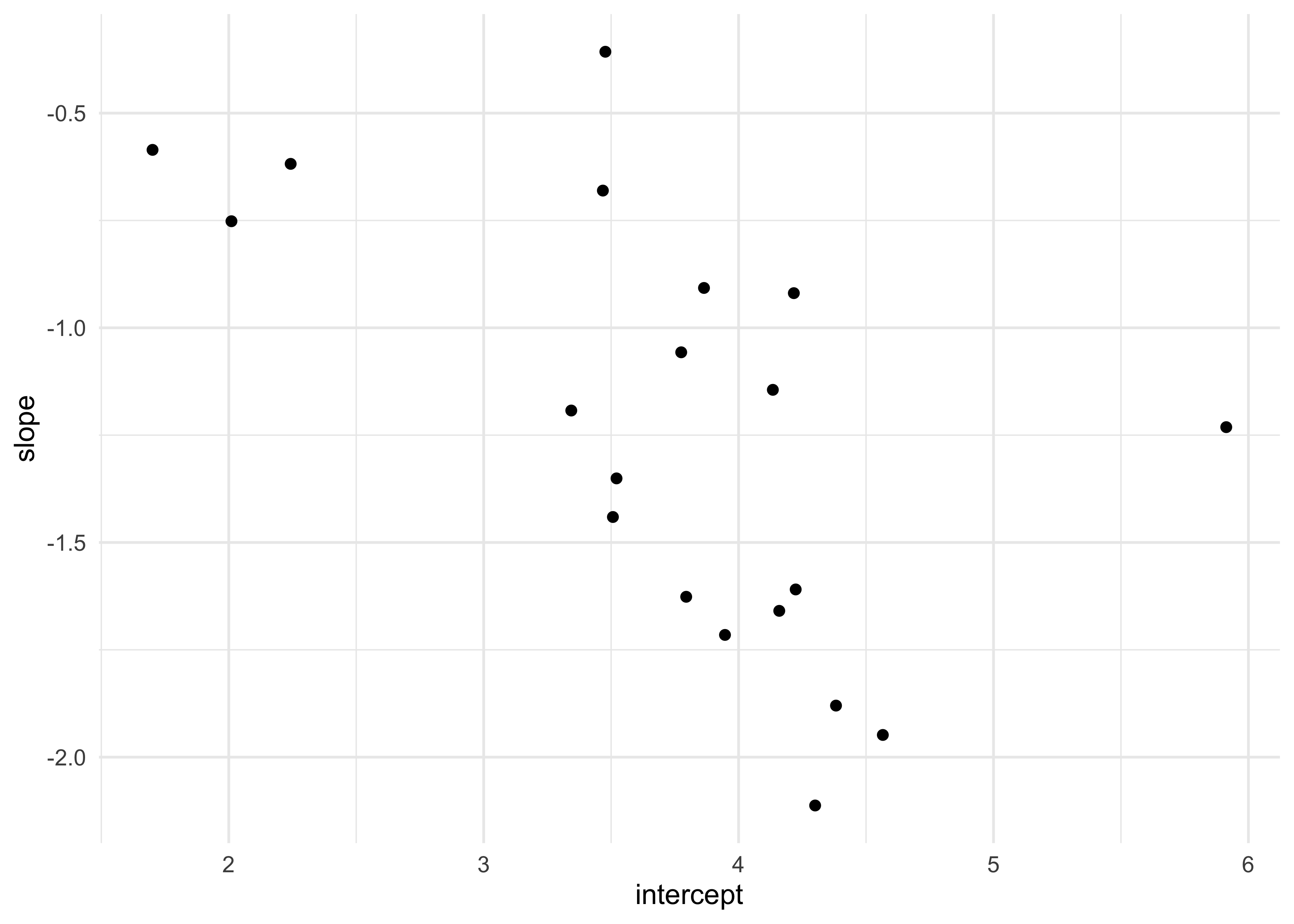
cor(a_cafe, b_cafe)
#> [1] -0.5721537
- plot of the varying effects
as.data.frame(vary_effects) %>%
as_tibble() %>%
set_names(c("intercept", "slope")) %>%
ggplot(aes(x = intercept, y = slope)) +
geom_point()
13.1.2 Simulate observations
- simulate the visits to each cafe
- 10 visits to each cafe, 5 in the morning and 5 in the afternoon
N_visits <- 10
afternoon <- rep(0:1, N_visits * N_cafes / 2)
cafe_id <- rep(1:N_cafes, each = N_visits)
# Get the average wait time for each cafe in the morning and afternoon.
mu <- a_cafe[cafe_id] + b_cafe[cafe_id] * afternoon
sigma <- 0.5 # Standard deviation within cafes
# Sample wait times with each cafes unique average wait time per time of day.
wait_times <- rnorm(N_visits*N_cafes, mu, sigma)
d <- tibble(cafe = cafe_id, afternoon, wait = wait_times)
d
#> # A tibble: 200 x 3
#> cafe afternoon wait
#> <int> <int> <dbl>
#> 1 1 0 5.00
#> 2 1 1 2.21
#> 3 1 0 4.19
#> 4 1 1 3.56
#> 5 1 0 4.00
#> 6 1 1 2.90
#> 7 1 0 3.78
#> 8 1 1 2.38
#> 9 1 0 3.86
#> 10 1 1 2.58
#> # … with 190 more rows
d %>%
ggplot(aes(x = wait, y = afternoon, color = factor(cafe))) +
geom_jitter(width = 0, height = 0.2) +
scale_color_manual(values = randomcoloR::distinctColorPalette(N_cafes),
guide = FALSE)
d %>%
ggplot(aes(x = wait, y = factor(cafe), color = factor(afternoon))) +
geom_point() +
scale_color_manual(values = c(blue, red))
13.1.3 The varying slopes model
- model with varying intercepts and slopes (explanation follows)
$$ W_i (_i, ) $$ $$
*i = *{$$i$$} + _{$$i$$} A_i $$ $$
(
, ) $$ $$
=
$$ $$
(0,10) $$ $$ (0,10) \ (0, 1) \ *(0, 1) \ *(0, 1) \ (2) $$
- the third like defines the population of varying intercepts and
slopes
- each cafe has an intercept and slope with a prior distribution defined by the 2D Gaussian distribution with means $\alpha$ and $\beta$ and covariance matrix $\text{S}$
$$ \begin{bmatrix} \alpha_\text{cafe} $$ $$ \beta_\text{cafe} \end{bmatrix} \sim \text{MVNormal}( \begin{bmatrix} \alpha $$ $$ \beta \end{bmatrix}, \textbf{S} ) $$
- the next line defines the variance-covariance matrix $\textbf{S}$
- factoring it into simple standard deviations $\sigma_\alpha$ and $\sigma_\beta$ and a correlation matrix $\textbf{R}$
- there are other ways to do this, but this formulation helps understand the inferred structure of the varying effects
$$ \textbf{S} = \begin{pmatrix} \sigma_\alpha & 0 $$ $$ 0 &\sigma_\beta \end{pmatrix} \textbf{R} \begin{pmatrix} \sigma_\alpha & 0 $$ $$ 0 &\sigma_\beta \end{pmatrix} $$
- the correlation matrix has a prior defined as
$\textbf{R} \sim \text{LKJcorr}(2)$
- the correlation matrix will have the structure: $\begin{pmatrix} 1 & \rho $$ $$ \rho & 1 \end{pmatrix}$ where $\rho$ is the correlation between the intercepts and slopes
- with additional varying slopes, there are more correlation parameters, but the $\text{LKJcorr}$ prior will still work
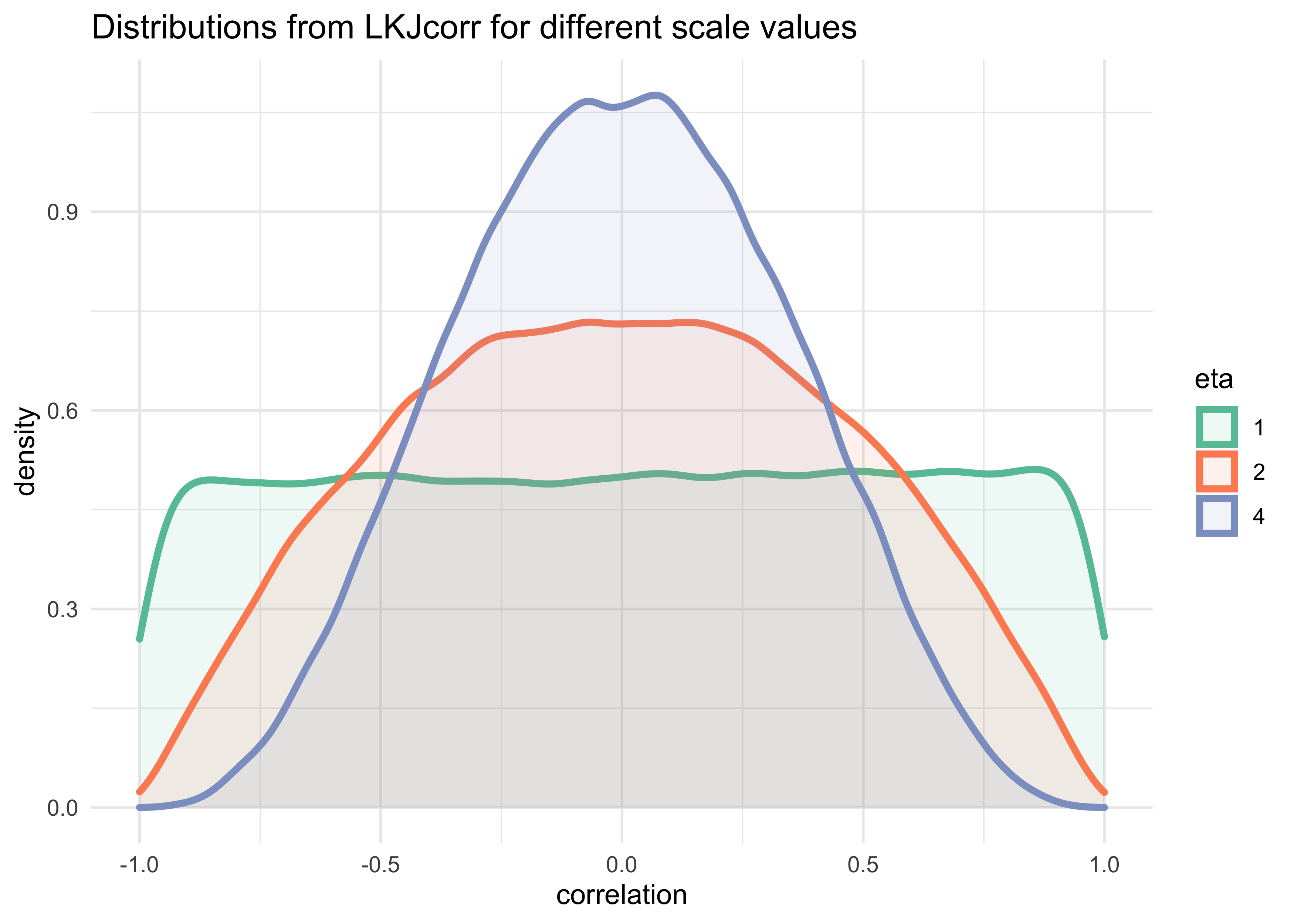
- the $\text{LKJcorr}(2)$ prior defines a weakly informative prior on $\rho$ that is skeptical of extreme correlations near -1 and 1
- it has a single parameter $\eta$ that controls how “skeptical”
the prior is of large correlations
- if $\eta=1$, the prior is flat from -1 to 1
- a large value of $\eta$ the mass of the distribution moves towards 0
tibble(eta = c(1, 2, 4)) %>%
mutate(value = map(eta, ~ rlkjcorr(1e5, K = 2, eta = .x)[, 1, 2])) %>%
unnest(value) %>%
ggplot(aes(x = value)) +
geom_density(aes(color = factor(eta), fill = factor(eta)),
size = 1.3, alpha = 0.1) +
scale_color_brewer(palette = "Set2") +
scale_fill_brewer(palette = "Set2") +
labs(x = "correlation",
y = "density",
title = "Distributions from LKJcorr for different scale values",
color = "eta", fill = "eta")
- now can fit the model
stash("m13_1", {
m13_1 <- map2stan(
alist(
wait ~ dnorm(mu, sigma),
mu <- a_cafe[cafe] + b_cafe[cafe]*afternoon,
c(a_cafe, b_cafe)[cafe] ~ dmvnorm2(c(a, b), sigma_cafe, Rho),
a ~ dnorm(0, 10),
b ~ dnorm(0, 10),
sigma_cafe ~ dcauchy(0, 2),
sigma ~ dcauchy(0, 2),
Rho ~ dlkjcorr(2)
),
data = d,
iter = 5e3, warmup = 2e3, chains = 2
)
})
#> Loading stashed object.
precis(m13_1, depth = 1)
#> 46 vector or matrix parameters hidden. Use depth=2 to show them.
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a 3.7411232 0.22805989 3.386143 4.1040327 6673.815 0.9998308
#> b -1.2443621 0.13662408 -1.458389 -1.0304374 5965.979 0.9999912
#> sigma 0.4648216 0.02624559 0.424598 0.5085003 6410.601 0.9999077
- inspection of the posterior distribution of varying effects
- start with the posterior correlation between intercepts and
slopes
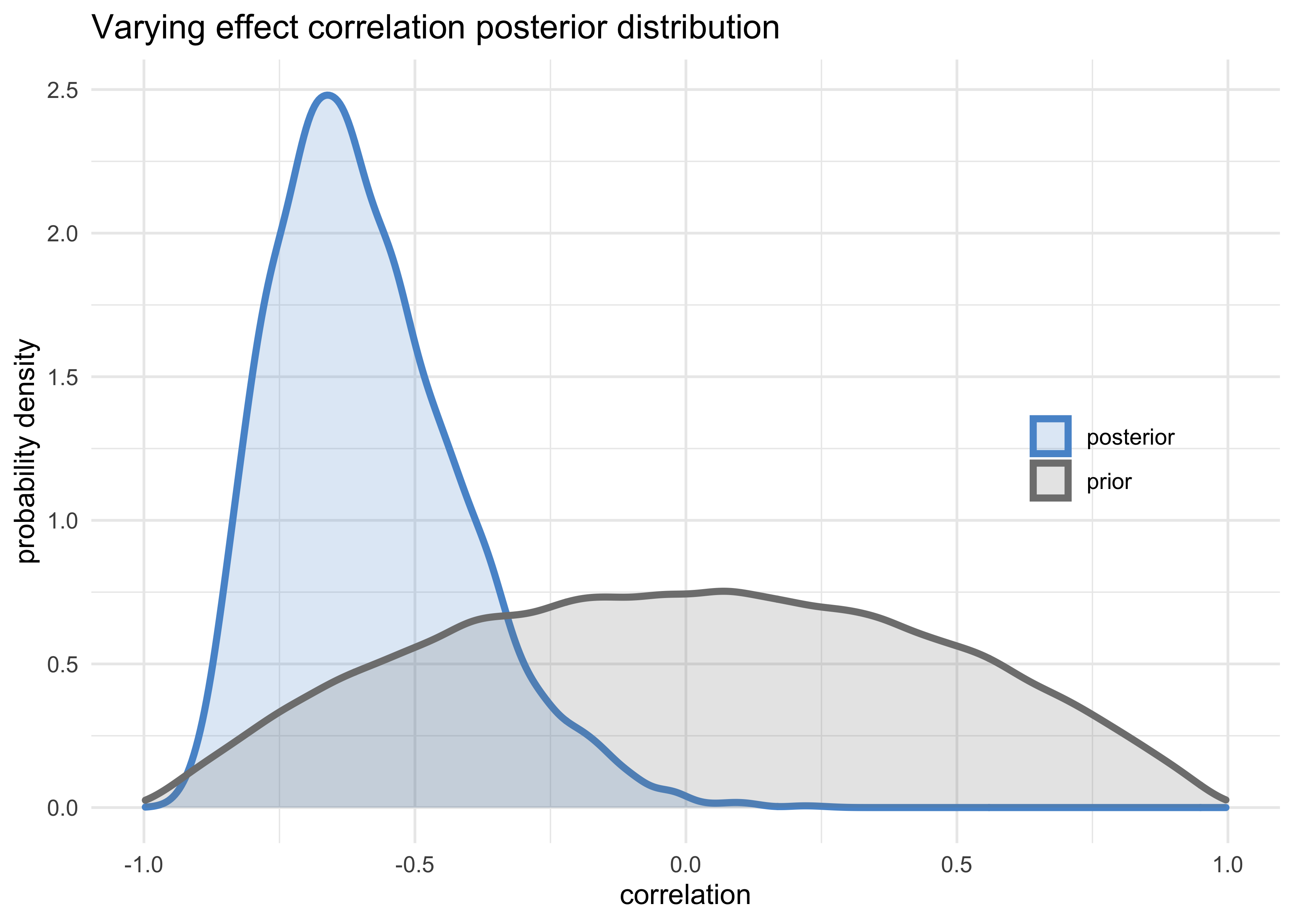
- the posterior distribution of the correlation between varying effects is decidedly negative
- start with the posterior correlation between intercepts and
slopes
post <- extract.samples(m13_1)
tribble(
~name, ~value,
"posterior", post$Rho[, 1, 2],
"prior", rlkjcorr(1e5, K = 2, eta = 2)[, 1, 2]
) %>%
unnest(value) %>%
ggplot(aes(x = value, color = name, fill = name)) +
geom_density(size = 1.3, alpha = 0.2) +
scale_color_manual(values = c(blue, grey)) +
scale_fill_manual(values = c(blue, grey)) +
theme(legend.title = element_blank(),
legend.position = c(0.85, 0.5)) +
labs(x = "correlation",
y = "probability density",
title = "Varying effect correlation posterior distribution")
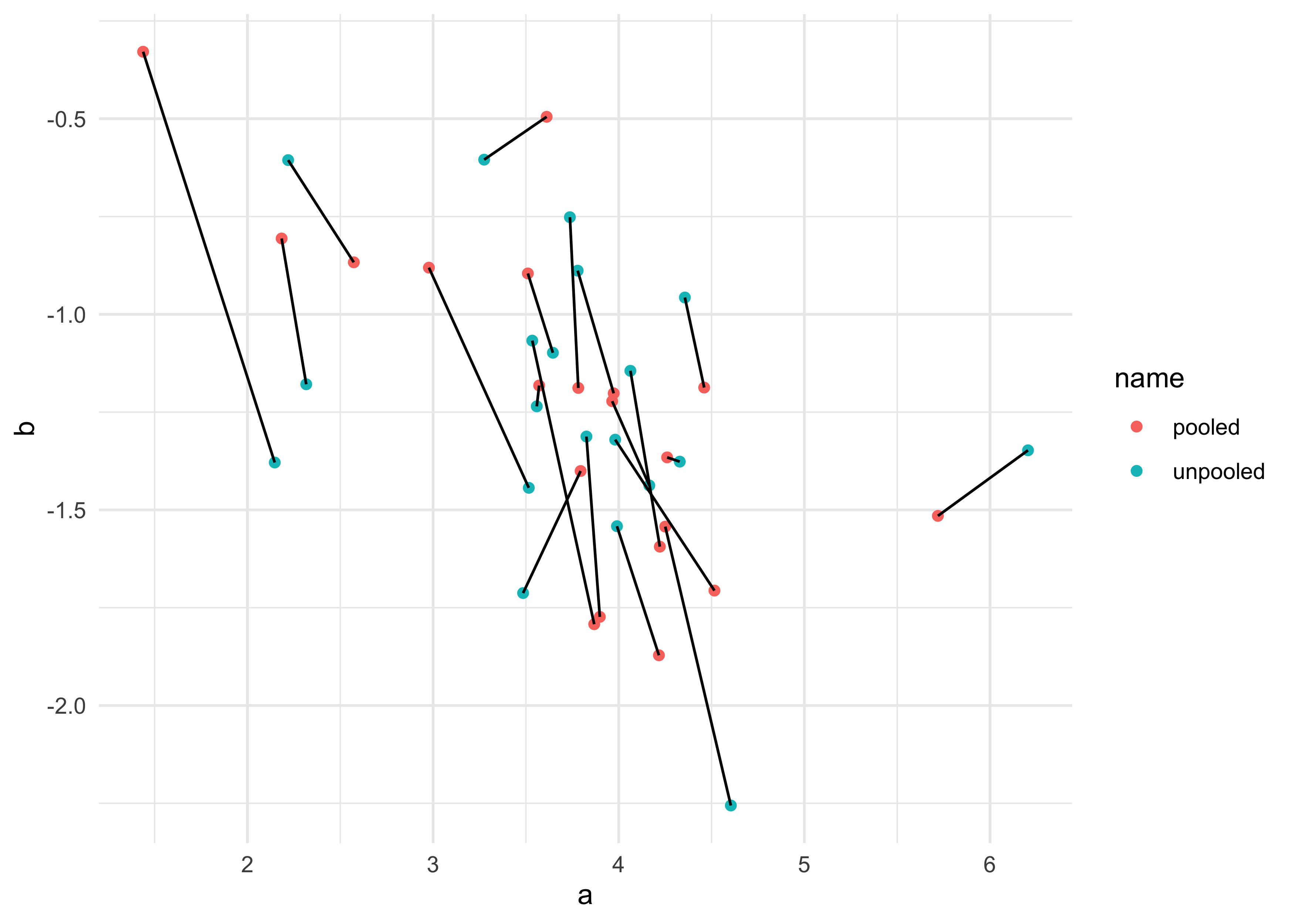
- consider the shrinkage
- the inferred correlation between varying effects pooled information across them
- and the inferred variation within each varying effect was pooled
- together the variances and correlation define a multivariate Gaussian prior for the varying effects
- this prior regularizes the intercepts and slopes
- plot the posterior mean varying effects
- compare them to the raw, unpooled estimates
- also plot the inferred prior for the population of intercepts and slopes
There is something wrong with the following 2 plots, but I cannot figure out what went wrong.
# Raw, unpooled estimates for alpha and beta.
a1 <- map_dbl(1:N_cafes, function(i) {
mean(d$wait[d$cafe == i & d$afternoon == 0])
})
b1 <- map_dbl(1:N_cafes, function(i) {
mean(d$wait[d$cafe == i & d$afternoon == 1])
})
b1 <- b1 - a1
# Extract posterior means of partially pooled estimates.
post <- extract.samples(m13_1)
a2 <- apply(post$a_cafe, 2, mean)
b2 <- apply(post$b_cafe, 2, mean)
tribble(
~ name, ~ a, ~ b,
"unpooled", a1, b1,
"pooled", a2, b2
) %>%
unnest(c(a, b)) %>%
group_by(name) %>%
mutate(cafe = row_number()) %>%
ungroup() %>%
ggplot(aes(x = a, y = b)) +
geom_point(aes(color = name)) +
geom_line(aes(group = cafe))
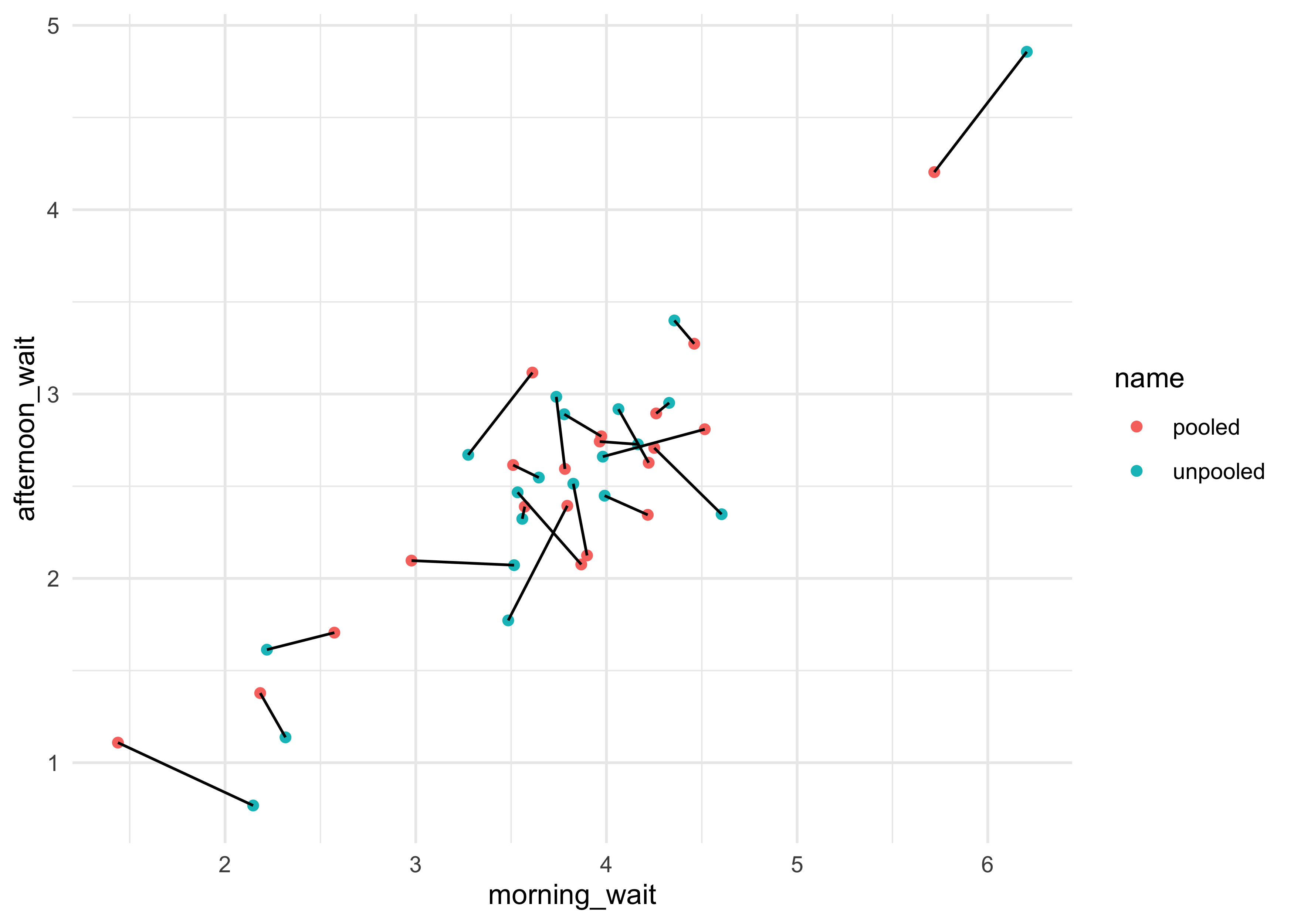
- can do the same for the estimated wait times for each cafe in the morning and afternoon
tribble(
~ name, ~ morning_wait, ~ afternoon_wait,
"unpooled", a1, a1 + b1,
"pooled", a2, a2 + b2
) %>%
unnest(c(morning_wait, afternoon_wait)) %>%
group_by(name) %>%
mutate(cafe = row_number()) %>%
ungroup() %>%
ggplot(aes(x = morning_wait, y = afternoon_wait)) +
geom_point(aes(color = name)) +
geom_line(aes(group = cafe))
13.2 Example: Admission decisions and gender
- return to the admissions data and use varying slopes
- help appreciate how variation in slopes arises
- and how correlation between intercepts and slopes can provide insight into the underlying process
- from previous models of the
UCBadmitdata:- important to have varying means across department otherwise, get wrong inference about gender
- did not account for variation in how departments treat male and female applications
data("UCBadmit")
d <- as_tibble(UCBadmit) %>%
janitor::clean_names() %>%
mutate(male = as.numeric(applicant_gender == "male"),
dept_id = coerce_index(dept))
13.2.1 Varying intercepts
- first model with only the varying intercepts
$$ A_i \sim \text{Binomial}(n_i, p_i) $$ $$ \text{logit}(p_i) = \alpha_{\text{dept}[i]} + \beta m_i $$ $$ \alpha_\text{dept} \sim \text{Normal}(\alpha, \sigma) $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta \sim \text{Normal}(0, 1) $$ $$ \sigma \sim \text{HalfCauchy}(0, 2) $$ $$ $$
stash("m13_2", {
m13_2 <- map2stan(
alist(
admit ~ dbinom(applications, p),
logit(p) <- a_dept[dept_id] + bm*male,
a_dept[dept_id] ~ dnorm(a, sigma_dept),
a ~ dnorm(0, 10),
bm ~ dnorm(0, 1),
sigma_dept ~ dcauchy(0, 2)
),
data = d,
warmup = 500, iter = 4500, chains = 3
)
})
#> Loading stashed object.
precis(m13_2, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_dept[1] 0.67633553 0.09912824 0.5172706 0.83647836 7434.742 0.9998820
#> a_dept[2] 0.62865617 0.11487786 0.4462060 0.81244427 7451.327 0.9999420
#> a_dept[3] -0.58407578 0.07381761 -0.7037524 -0.46666875 9691.393 1.0000113
#> a_dept[4] -0.61607540 0.08473505 -0.7513828 -0.47998152 8592.307 0.9998870
#> a_dept[5] -1.05883405 0.09832475 -1.2173306 -0.90286742 12118.564 0.9999736
#> a_dept[6] -2.60914320 0.15654731 -2.8607355 -2.36390139 10442.752 1.0000161
#> a -0.61042302 0.67864636 -1.6337291 0.40209932 5772.182 1.0004375
#> bm -0.09471168 0.08101324 -0.2257535 0.03522607 5929.314 1.0000314
#> sigma_dept 1.49095940 0.58217912 0.8576455 2.52329901 5681.029 1.0003271
- interpretation
- effect of male is similar that found in Chapter 10 (“Counting
and Classification”)
- the intercept is effectively uninteresting, if perhaps slightly negative
- because we included the global mean $\alpha$ in the prior for
the varying intercepts, the
a_dept[i]values are all deviations froma
- effect of male is similar that found in Chapter 10 (“Counting
and Classification”)
13.2.2 Varying effects of being male
- now we can consider the variation in gender bias among departments
- use varying slopes
- the data is imbalanced
- the sample sizes vary a lot across departments
- pooling will have a stronger effect for cases with fewer applicants
$$ A_i (n_i, p_i) $$ $$ (p_i) = *{$$i$$} + *{$$i$$} m_i \
(
, ) $$ $$
(0, 10) $$ $$ (0, 1) \
=
$$ $$
(*, *) (0, 2) $$ $$ (2) $$
stash("m13_3", {
m13_3 <- map2stan(
alist(
admit ~ dbinom(applications, p),
logit(p) <- a_dept[dept_id] + bm_dept[dept_id]*male,
c(a_dept, bm_dept)[dept_id] ~ dmvnorm2(c(a, bm), sigma_dept, Rho),
a ~ dnorm(0, 10),
bm ~ dnorm(0, 1),
sigma_dept ~ dcauchy(0, 2),
Rho ~ dlkjcorr(2)
),
data = d,
warmup = 1e3, iter = 5e3, chains = 4
)
})
#> Loading stashed object.
precis(m13_3, depth = 2)
#> 4 matrix parameters hidden. Use depth=3 to show them.
#> mean sd 5.5% 94.5% n_eff Rhat4
#> bm_dept[1] -0.79410911 0.26621581 -1.2314113 -0.3769640 7777.776 1.0001133
#> bm_dept[2] -0.21452957 0.32827223 -0.7442305 0.2976597 7706.047 1.0002087
#> bm_dept[3] 0.08210299 0.13916525 -0.1436567 0.3047949 14294.571 1.0001411
#> bm_dept[4] -0.09107295 0.14166835 -0.3189406 0.1369298 12245.517 1.0003159
#> bm_dept[5] 0.12616695 0.18522579 -0.1659927 0.4251783 13979.943 0.9998708
#> bm_dept[6] -0.12371601 0.26875987 -0.5601516 0.3021377 11343.429 1.0000722
#> a_dept[1] 1.30684485 0.25312427 0.9145892 1.7201724 7768.113 1.0000828
#> a_dept[2] 0.74419573 0.32704100 0.2349881 1.2721056 7974.643 1.0001940
#> a_dept[3] -0.64703547 0.08559338 -0.7820730 -0.5101126 15352.790 1.0001005
#> a_dept[4] -0.61945023 0.10575932 -0.7888304 -0.4512668 12336.691 1.0001413
#> a_dept[5] -1.13473049 0.11377576 -1.3178663 -0.9556382 14357.673 0.9999653
#> a_dept[6] -2.60003282 0.19884720 -2.9234768 -2.2882146 13404.431 1.0000262
#> a -0.50152540 0.73756490 -1.6147961 0.6207689 8686.590 1.0001169
#> bm -0.16333123 0.23571418 -0.5286623 0.1998200 8991.565 1.0000629
#> sigma_dept[1] 1.68379712 0.65077176 0.9809923 2.7911987 7190.902 1.0000942
#> sigma_dept[2] 0.50115741 0.25052608 0.2116896 0.9321738 7771.766 1.0003994
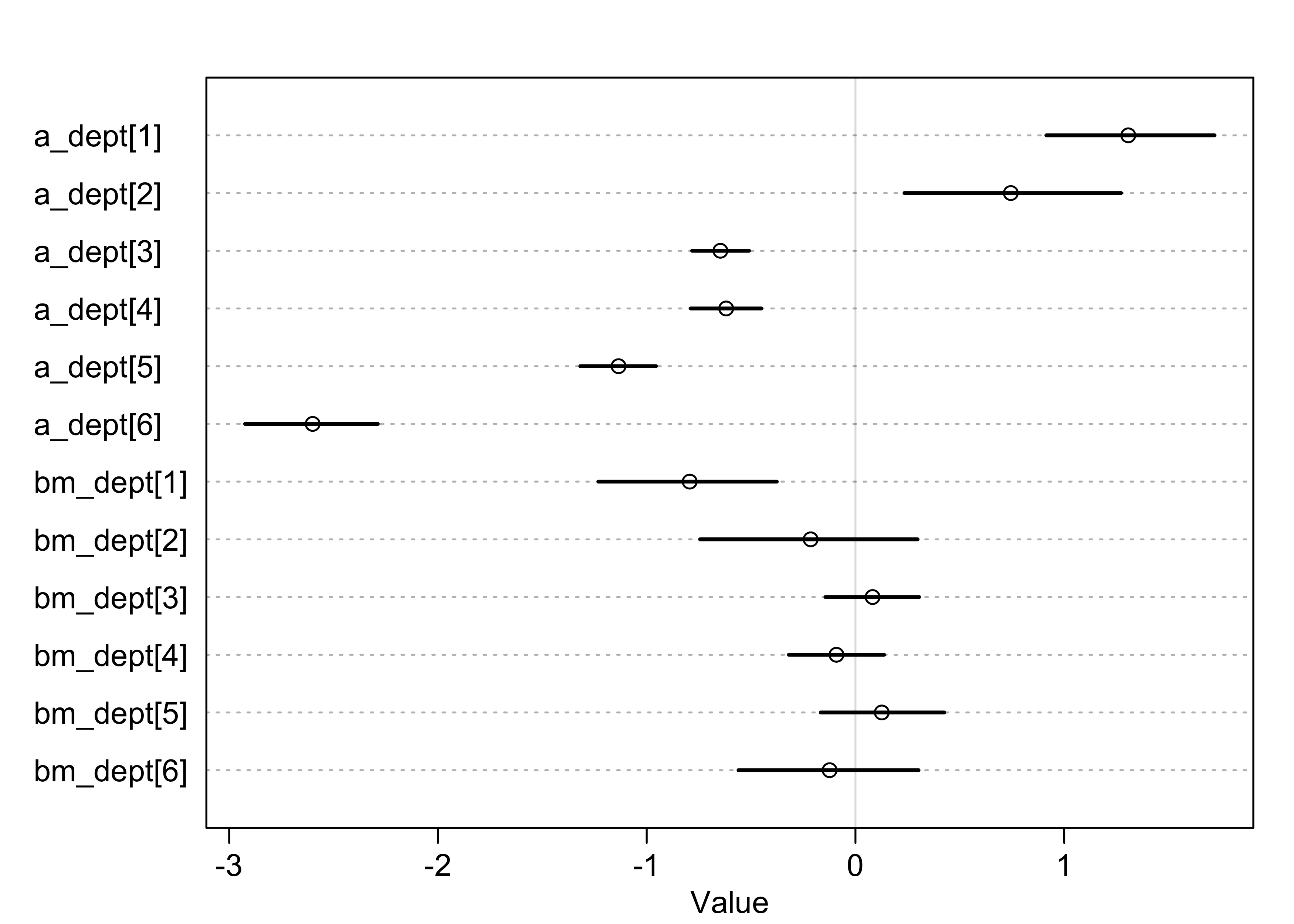
- focus on what the addition of varying slopes has revealed
- plot below shows marginal posterior distributions for the varying effects
- the intercepts are quite varied, but the slopes are all quite
close to 0
- suggests that the departments had different rates of admissions, but none discriminated between male and females
- one standout is the slope for department 1 which suggests
some bias against females
- department 1 also has the largest intercept, so look into the correlation between slopes and intercepts next
plot(precis(m13_3, pars = c("a_dept", "bm_dept"), depth = 2))
13.2.3 Shrinkage
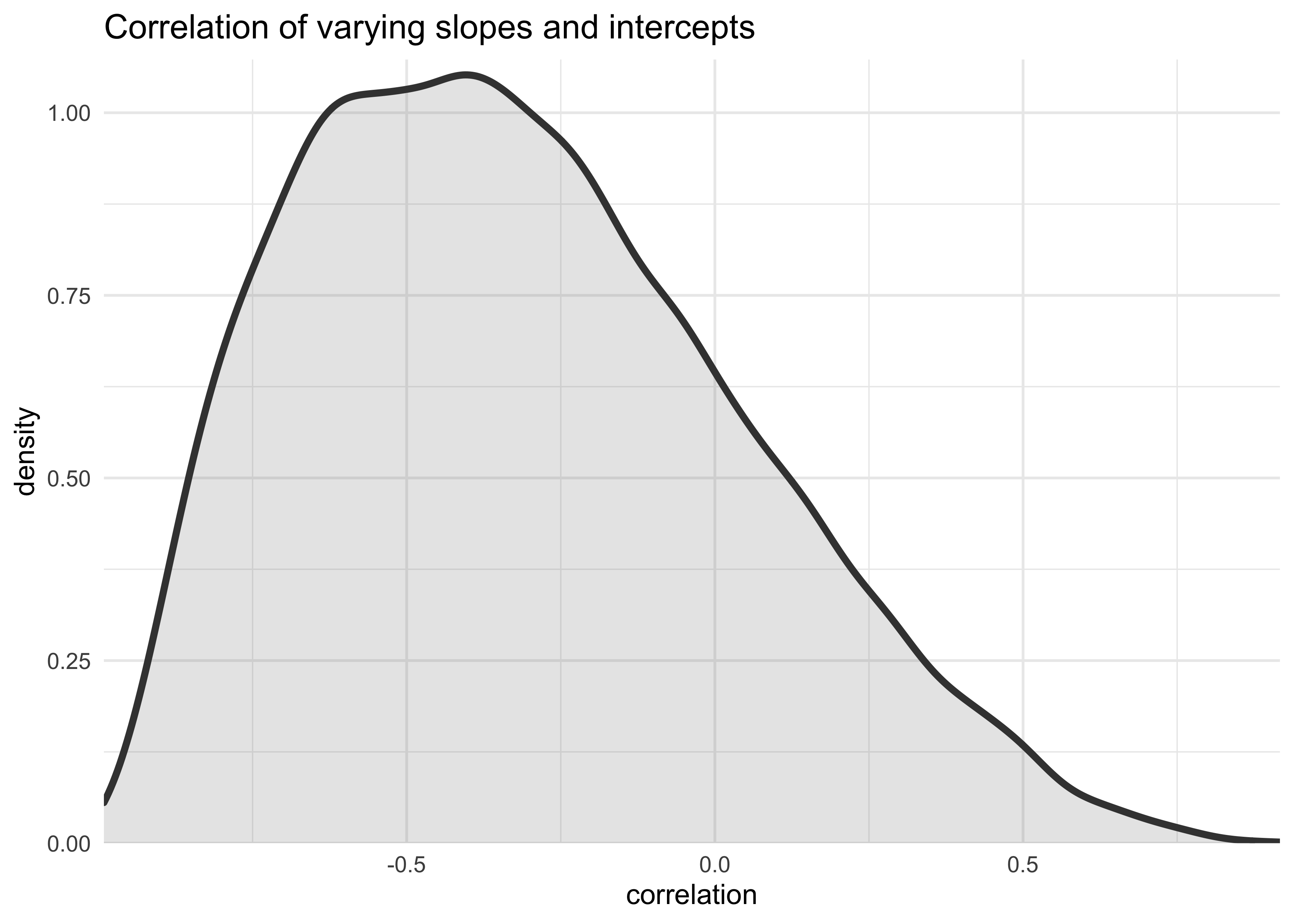
- following plot shows the posterior distribution for the correlation
between slope and intercept
- negative correlation: the higher the admissions rate, the lower the slope
post <- extract.samples(m13_3)
tibble(posterior_rho = post$Rho[, 1, 2]) %>%
ggplot(aes(x = posterior_rho)) +
geom_density(size = 1.3, color = dark_grey, fill = grey, alpha = 0.2) +
scale_x_continuous(expand = expansion(mult = c(0, 0))) +
scale_y_continuous(expand = expansion(mult = c(0, 0.02))) +
labs(x = "correlation",
y = "density",
title = "Correlation of varying slopes and intercepts")
13.2.4 Model comparison
- also fit a model that ignores gender for purposes of comparison
stash("m13_4", {
m13_4 <- map2stan(
alist(
admit ~ dbinom(applications, p),
logit(p) <- a_dept[dept_id],
a_dept[dept_id] ~ dnorm(a, sigma_dept),
a ~ dnorm(0, 10),
sigma_dept ~ dcauchy(0, 2)
),
data = d,
warmup = 500, iter = 4500, chains = 3
)
})
#> Loading stashed object.
compare(m13_2, m13_3, m13_4)
#> WAIC SE dWAIC dSE pWAIC weight
#> m13_3 5190.940 57.26937 0.00000 NA 11.094350 0.987737123
#> m13_4 5200.945 56.84874 10.00469 6.820917 5.939283 0.006639735
#> m13_2 5201.277 56.93248 10.33705 6.516639 6.864329 0.005623142
- interpretation:
- the model with no slope for differences in gender
m13_4performs the same out-of-sample performance as the model with a single slope for a constant effect of genderm13_2 - the model with varying slopes suggests that even though the slope is near zero, it is worth modeling as a separate distribution
- the model with no slope for differences in gender
13.3 Example: Cross-classified chimpanzees with varying slopes
- use chimpanzee data to model multiple varying intercepts and/or
slopes
- varying intercepts for
actorandblock - varying slopes for prosocial option and the interaction between prosocial and the presence of another chimpanzee
- varying intercepts for
- non-centered parameterization (see later for explanation and
example)
- there are always several ways to formulate a model that are mathematically equivalent
- however, they can result in different sampling results, so the parameterization is part of the model
- cross-classified varying slopes model
- use multiple linear models to compartmentalize sub-models for the intercepts and each slope
- $\mathcal{A}i$, $\mathcal{B}{P,i}$, and $\mathcal{B}_{PC,i}$ are the sub-models
$$ L_i \sim \text{Binomial}(1, p_i) $$ $$ \text{logit}(p_i) = \mathcal{A}i + (\mathcal{B}{p,i} + \mathcal{B}{PC,i} C_i) P_i $$ $$ \mathcal{A}i = \alpha + \alpha{\text{actor}[i]} + \alpha{\text{block}[i]} $$ $$ \mathcal{B}{P,i} = \beta_P + \beta{P,\text{actor}[i]} + \beta_{P,\text{block}[i]} $$ $$ \mathcal{B}_{PC,i} = \beta_P + \beta_{PC,\text{actor}[i]} + \beta_{PC,\text{block}[i]} $$ $$ $$
- below is the formulation for the multivariate priors
- one multivariate Gaussian per cluster of the data (
actorandblock) - for this model, each is 3D, one for each variable in the model
- this can be adjusted to have different varying effects in different cluster types
- these priors state that the actors and blocks come from
different statistical populations
- within each, three features for each actor or block are related through a covariance matrix for the population ($\textbf{S}$)
- the mean for each prior is 0 because there is an average value in the linear models already ($\alpha$, $\beta_P$, and $\beta_{PC,i}$)
- one multivariate Gaussian per cluster of the data (
$$
$$ $$
$$
data("chimpanzees")
d <- as_tibble(chimpanzees) %>%
select(-recipient) %>%
rename(block_id = block)
stash("m13_6", {
m13_6 <- map2stan(
alist(
pulled_left ~ dbinom(1, p),
logit(p) <- A + (BP + BPC*condition) * prosoc_left,
A <- a + a_actor[actor] + a_block[block_id],
BP <- bp + bp_actor[actor] + bp_block[block_id],
BPC <- bpc + bpc_actor[actor] + bpc_block[block_id],
c(a_actor, bp_actor, bpc_actor)[actor] ~ dmvnorm2(0, sigma_actor, Rho_actor),
c(a_block, bp_block, bpc_block)[block_id] ~ dmvnorm2(0, sigma_block, Rho_block),
c(a, bp, bpc) ~ dnorm(0, 1),
sigma_actor ~ dcauchy(0, 2),
sigma_block ~ dcauchy(0, 2),
Rho_actor ~ dlkjcorr(4),
Rho_block ~ dlkjcorr(4)
),
data = d,
iter = 5e3, warmup = 1e3, chains = 3
)
})
#> Loading stashed object.
precis(m13_6, depth = 1)
#> 63 vector or matrix parameters hidden. Use depth=2 to show them.
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a 0.23573453 0.6666099 -0.90821904 1.2611100 934.458 1.001440
#> bp 0.70892674 0.4052890 0.05423901 1.3312395 3817.263 1.000350
#> bpc -0.03931986 0.4317621 -0.68617116 0.6610651 2216.997 1.001548
- there was an issue with the HMC sampling
- can often just do more sampling to get over it, but other times the chains may not converge
- this is where non-centered parameterization can help
- In map2stan(alist(pulled_left ~ dbinom(1, p), logit(p) <- A + (BP +
- There were 559 divergent iterations during sampling. Check the chains (trace plots, n_eff, Rhat) carefully to ensure they are valid.
- use non-centered parameterization to help with potentially
diverging chains
- use an alternative parameterization of the model using
dmvnormNC() - mathematically equivalent to the first
- use an alternative parameterization of the model using
stash("m13_6nc", {
m13_6nc <- map2stan(
alist(
pulled_left ~ dbinom(1, p),
logit(p) <- A + (BP + BPC*condition) * prosoc_left,
A <- a + a_actor[actor] + a_block[block_id],
BP <- bp + bp_actor[actor] + bp_block[block_id],
BPC <- bpc + bpc_actor[actor] + bpc_block[block_id],
c(a_actor, bp_actor, bpc_actor)[actor] ~ dmvnormNC(sigma_actor, Rho_actor),
c(a_block, bp_block, bpc_block)[block_id] ~ dmvnormNC(sigma_block, Rho_block),
c(a, bp, bpc) ~ dnorm(0, 1),
sigma_actor ~ dcauchy(0, 2),
sigma_block ~ dcauchy(0, 2),
Rho_actor ~ dlkjcorr(4),
Rho_block ~ dlkjcorr(4)
),
data = d,
iter = 5e3, warmup = 1e3, chains = 3
)
})
#> Loading stashed object.
precis(m13_6nc, depth = 1)
#> 120 vector or matrix parameters hidden. Use depth=2 to show them.
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a 0.25351038 0.6573804 -0.7999981 1.2938998 4714.341 1.0005064
#> bp 0.71776740 0.4010091 0.0716376 1.3317852 10602.170 1.0003175
#> bpc -0.02295208 0.4303382 -0.6776100 0.6685359 11206.151 0.9999246
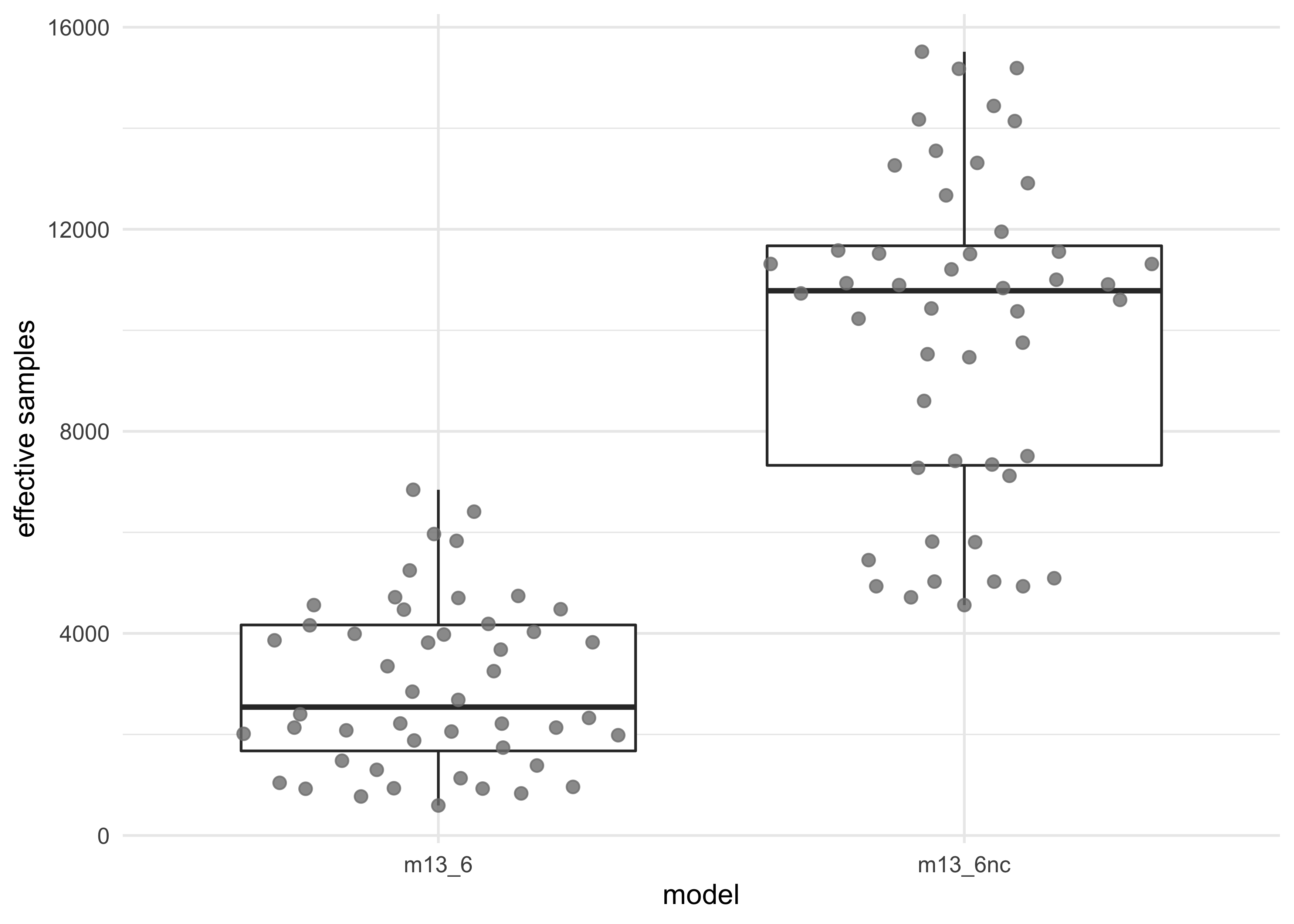
- the non-centered parameterization helped by sampling faster and more effectively
get_neff <- function(mdl, depth) {
x <- precis(mdl, depth = depth)
neff_idx <- which(x@names == "n_eff")
x@.Data[neff_idx]
}
tibble(m13_6 = get_neff(m13_6, depth = 2),
m13_6nc = get_neff(m13_6nc, depth = 2)) %>%
pivot_longer(tidyselect::everything()) %>%
unnest(value) %>%
ggplot(aes(name, value)) +
geom_boxplot(outlier.shape = NA) +
ggbeeswarm::geom_quasirandom(color = grey, size = 2, alpha = 0.8) +
labs(x = "model",
y = "effective samples")
#> 18 matrix parameters hidden. Use depth=3 to show them.
#> 75 matrix parameters hidden. Use depth=3 to show them.
- can see the number of effective parameters is much smaller than the real number
WAIC(m13_6nc)
#> WAIC lppd penalty std_err
#> 1 534.7752 -249.032 18.35563 19.88306
- the standard deviation parameters of the random effects provides a
measure of how much regularization was applied
- the first index for each sigma is the varying intercept standard deviation while the other two are the slopes
- the values are pretty small suggesting there was a good amount of shrinkage
precis(m13_6nc, depth = 2, pars = c("sigma_actor", "sigma_block"))
#> mean sd 5.5% 94.5% n_eff Rhat4
#> sigma_actor[1] 2.3327748 0.9056939 1.31285970 3.952849 5816.898 1.000055
#> sigma_actor[2] 0.4537137 0.3627789 0.04461175 1.098381 7510.460 1.000236
#> sigma_actor[3] 0.5205115 0.4761865 0.03924063 1.385698 7414.251 1.000002
#> sigma_block[1] 0.2295740 0.2077322 0.01855211 0.598803 7343.314 1.000648
#> sigma_block[2] 0.5716266 0.4057379 0.07362251 1.274700 4560.184 1.000127
#> sigma_block[3] 0.5032187 0.4180730 0.04109647 1.243568 7119.573 1.000062
- compare the varying slopes model to the simpler varying intercepts
model from the previous chapter
- the results indicate that there isn’t too much difference between the two models
- meaning there isn’t much difference in the slopes between
actornorblock
stash("m12_5", {
m12_5 <- map2stan(
alist(
pulled_left ~ dbinom(1, p),
logit(p) <- a + a_actor[actor] + a_block[block_id] + (bp + bpc*condition)*prosoc_left,
a_actor[actor] ~ dnorm(0, sigma_actor),
a_block[block_id] ~ dnorm(0, sigma_block),
a ~ dnorm(0, 10),
bp ~ dnorm(0, 10),
bpc ~ dnorm(0, 10),
sigma_actor ~ dcauchy(0, 1),
sigma_block ~ dcauchy(0, 1)
),
data = d,
warmup = 1e3,
iter = 6e3,
chains = 4
)
})
#> Loading stashed object.
compare(m13_6nc, m12_5)
#> WAIC SE dWAIC dSE pWAIC weight
#> m12_5 532.4494 19.66977 0.0000 NA 10.29711 0.7618593
#> m13_6nc 534.7752 19.88306 2.3258 4.070507 18.35563 0.2381407
13.4 Continuous categories and the Gaussian process
- so far, all varying intercepts and slopes were defined over
discrete, unordered categories
- now learn how to use continuous dimensions of variation
- e.g.: age, income, social standing
- now learn how to use continuous dimensions of variation
- Gaussian process regression: method for applying a varying effect
to continuous categories
- estimates a unique intercept/slope for any value in the variable and applies shrinkage to these values
- simple outline of the process:
- calculate differences between all data points in the category
- the model estimates a function for the covariance between pairs of cases at each distance
- the coviariance function is the generalization of the varying effects approach to continuous categories
13.4.1 Example: Spatial autocorrelation in Oceanic tools
- in previous modeling of the Oceanic societies data, used a binary
contact predictor
- want to make a model that keeps this as a continuous variable
- many reasons why islands near each other would have similar tools
- the process:
- define a distance matrix among the societies
- then estimate how similarity in tool counts depends on geographic distance
data("islandsDistMatrix")
Dmat <- islandsDistMatrix
colnames(Dmat) <- c("Ml","Tiv","SC","Ya","Fi","Tr","Ch","Mn","To","Ha")
round(Dmat, 1)
#> Ml Tiv SC Ya Fi Tr Ch Mn To Ha
#> Malekula 0.0 0.5 0.6 4.4 1.2 2.0 3.2 2.8 1.9 5.7
#> Tikopia 0.5 0.0 0.3 4.2 1.2 2.0 2.9 2.7 2.0 5.3
#> Santa Cruz 0.6 0.3 0.0 3.9 1.6 1.7 2.6 2.4 2.3 5.4
#> Yap 4.4 4.2 3.9 0.0 5.4 2.5 1.6 1.6 6.1 7.2
#> Lau Fiji 1.2 1.2 1.6 5.4 0.0 3.2 4.0 3.9 0.8 4.9
#> Trobriand 2.0 2.0 1.7 2.5 3.2 0.0 1.8 0.8 3.9 6.7
#> Chuuk 3.2 2.9 2.6 1.6 4.0 1.8 0.0 1.2 4.8 5.8
#> Manus 2.8 2.7 2.4 1.6 3.9 0.8 1.2 0.0 4.6 6.7
#> Tonga 1.9 2.0 2.3 6.1 0.8 3.9 4.8 4.6 0.0 5.0
#> Hawaii 5.7 5.3 5.4 7.2 4.9 6.7 5.8 6.7 5.0 0.0
- the likelihood and linear model for this model look the same as
before:
- Poisson likelihood with a varying intercept linear model with a log link function
- the $\gamma_{\text{society}}$ is the varying intercept
- regular coefficient for log population
- determine if accounting for spatial similarity will wash out the association between log population and total number of tools
$$ T_i \sim \text{Poisson}(\lambda_i) $$ $$ \log \lambda_i = \alpha + \gamma_{\text{society}[i]} + \beta_P \log P_i $$
- add in a multivariate prior for the intercepts for the Gaussian
process
- first is the 10-dimensional Gaussian prior for the intercepts
- $\textbf{K}$ is the covariance matrix between any pairs of
societies $i$ and $j$
- three new parameters: $\eta$, $\rho$, and $\sigma$
- the Gaussian shape comes from $\exp(-\rho^2 D_{ij}^2)$
where $D_{ij}$ is the distance between societies $i$ and
$j$
- says that the covariance between two societies declines exponentially with the squared distance
- $\rho$ determines the rate of decline (large = declines rapidly with distance)
- the distance need not be squared, but usually is because it is often a more realistic model and fits more easily
- $\eta^2$ is the maximum covariance between two societies $i$ and $j$
- $\delta_{ij}\sigma^2$ provides for extra covariance beyond
$\eta^2$ when $i=j$
- the function $\delta_{ij}$ is 1 when $i=j$, else 0
- this only matters if there is more than one data point per group (which there isn’t in the Oceanic example)
- therefore, $sigma$ describes how the observations for a single category covary
$$ \gamma \sim \text{MVNormal}([0, …, 0], \textbf{K}) $$ $$ \textbf{K}{ij} = \eta^2 \exp(-\rho^2 D{ij}^2) + \delta_{ij}\sigma^2 $$
- the full model
- create priors for $\eta^2$ and $\rho^2$ because is easier to fit
- set $\sigma$ as a small constant because it does not get used in this model (see above)
$$ T_i \sim \text{Poisson}(\lambda_i) $$ $$ \log \lambda_i = \alpha + \gamma_{\text{society}[i]} + \beta_P \log P_i $$ $$ \gamma \sim \text{MVNormal}([0, …, 0], \textbf{K}) $$ $$ \textbf{K}_{ij} = \eta^2 \exp(-\rho^2 D_{ij}^2) + \delta_{ij}(0.01) $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta_P \sim \text{Normal}(0, 1) $$ $$ \eta^2 \sim \text{HalfCauchy}(0, 1) $$ $$ \rho^2 \sim \text{HalfCauchy}(0, 1) $$
- fit using
map2stan()- use
GPL2()in order to use a squared distance Gaussian process prior
- use
data("Kline2")
d <- as_tibble(Kline2) %>%
mutate(society = row_number())
stash("m13_7", {
m13_7 <- map2stan(
alist(
total_tools ~ dpois(lambda),
log(lambda) <- a + g[society] + bp*logpop,
g[society] ~ GPL2(Dmat, etasq, rhosq, 0.01),
a ~ dnorm(0, 10),
bp ~ dnorm(0, 1),
etasq ~ dcauchy(0, 1),
rhosq ~ dcauchy(0, 1)
),
data = list(total_tools = d$total_tools,
logpop = d$logpop,
society = d$society,
Dmat = islandsDistMatrix),
warmup = 2e3, iter = 1e4, chains = 4
)
})
#> Loading stashed object.
plot(m13_7)
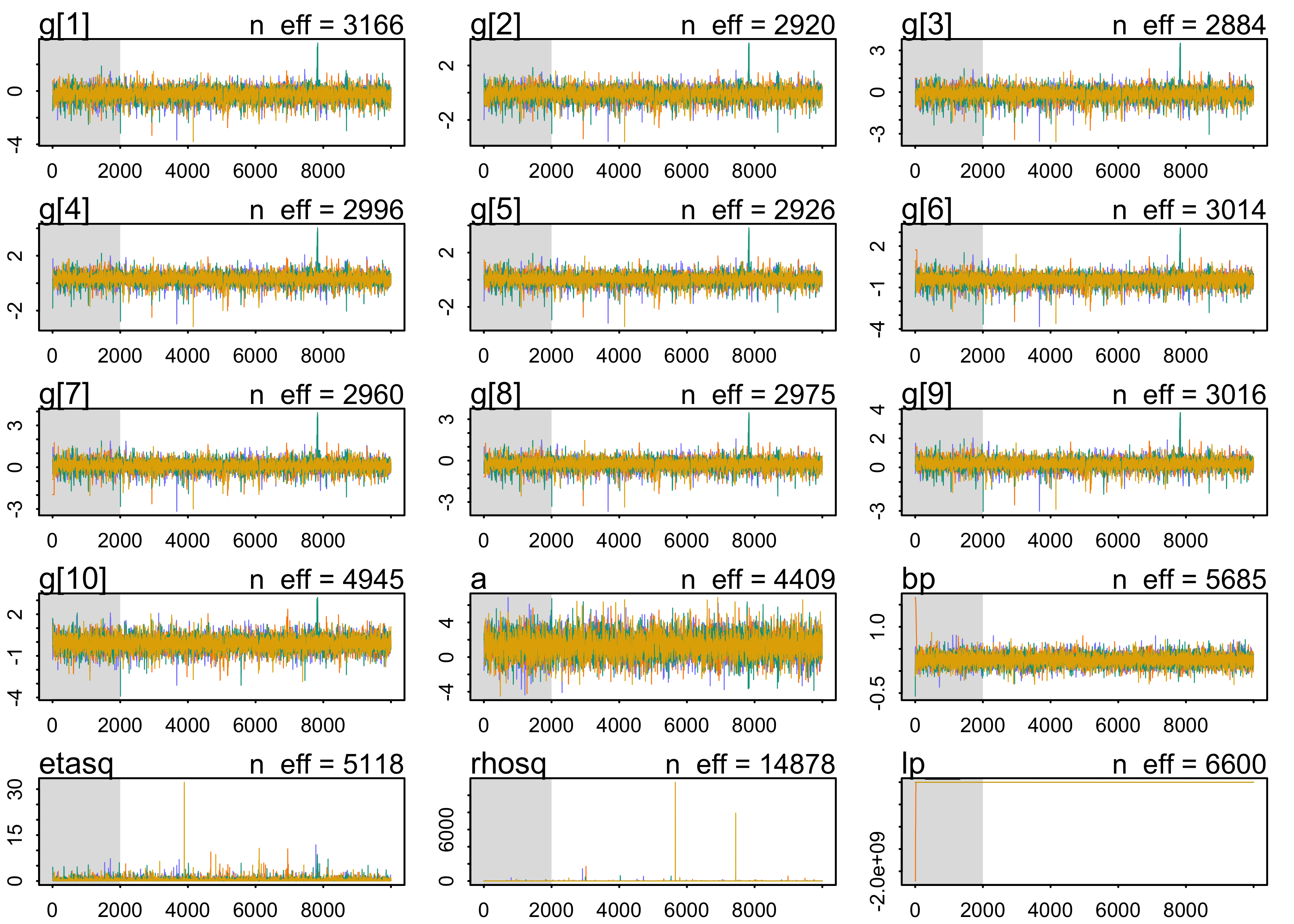
precis(m13_7, depth=2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> g[1] -0.27856102 0.4398912 -0.99830940 0.34423072 3166.290 1.000329
#> g[2] -0.13030314 0.4275290 -0.82517622 0.48309679 2920.299 1.000473
#> g[3] -0.17472739 0.4129535 -0.85615406 0.39565329 2883.929 1.000473
#> g[4] 0.29041188 0.3673727 -0.27347655 0.81883438 2996.021 1.000574
#> g[5] 0.01904426 0.3632424 -0.54846932 0.52312820 2925.635 1.000645
#> g[6] -0.46551126 0.3747591 -1.07984663 0.02455556 3013.954 1.000428
#> g[7] 0.08850494 0.3588191 -0.47706011 0.58111284 2959.900 1.000635
#> g[8] -0.27052427 0.3616190 -0.84554239 0.21383134 2974.716 1.000614
#> g[9] 0.22699085 0.3398043 -0.28673166 0.70034159 3015.712 1.000794
#> g[10] -0.12811344 0.4518739 -0.83076660 0.55536172 4945.226 1.001099
#> a 1.30857523 1.1568335 -0.49441544 3.17347739 4408.741 1.000326
#> bp 0.24645057 0.1140199 0.06828176 0.42625222 5685.467 1.000516
#> etasq 0.33740391 0.5130216 0.04065684 0.99697571 5117.732 1.000657
#> rhosq 2.81525694 93.1286058 0.05231803 3.80800354 14877.678 1.000069
- interpretation:
- the coefficient for log population
bpis the same as before adding in the Gaussian process for varying intercepts- the association between tool counts and population cannot be explained by spatial correlations
- the coefficient for log population
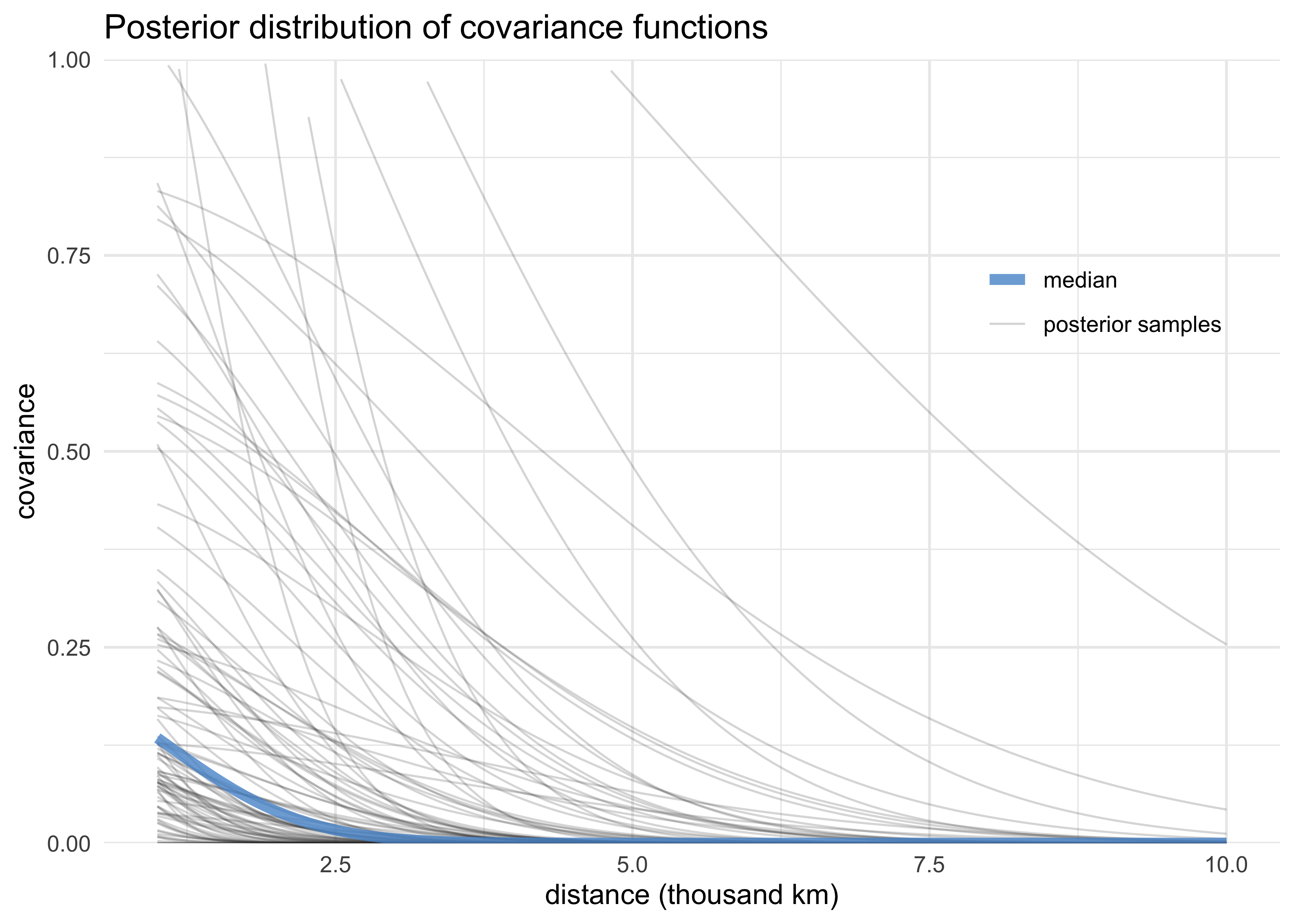
- plot posterior of covariance functions using
rhospandetasqsamples
post <- extract.samples(m13_7)
median_covar_df <- tibble(etasq = median(post$etasq),
rhosq = median(post$rhosq),
distance = seq(1, 10, length.out = 100)) %>%
mutate(density = etasq * exp(-rhosq * distance^2),
color = "median",
group = "median")
sample_covar_df <- tibble(etasq = post$etasq[1:100],
rhosq = post$rhosq[1:100]) %>%
mutate(group = as.character(row_number()),
distance = list(rep(seq(1, 10, length.out = 100), n()))) %>%
unnest(distance) %>%
mutate(density = etasq * exp(-rhosq * distance^2),
color = "posterior samples")
bind_rows(median_covar_df, sample_covar_df) %>%
ggplot(aes(distance, density)) +
geom_line(aes(group = group, alpha = color, size = color, color = color)) +
scale_alpha_manual(values = c(0.8, 0.2)) +
scale_size_manual(values = c(2, 0.4)) +
scale_color_manual(values = c(blue, "grey20")) +
scale_y_continuous(limits = c(0, 1),
expand = c(0, 0)) +
theme(legend.position = c(0.85, 0.7),
legend.title = element_blank()) +
labs(x = "distance (thousand km)",
y = "covariance",
title = "Posterior distribution of covariance functions")
#> Warning: Removed 11100 row(s) containing missing values (geom_path).
- consider the covariations among societies that are implied by the
posterior median
- first must pass the parameters back through the covariance matrix $\textbf{K}$
- then convert $\textbf{K}$ to a correlation matrix
Rho
# 1. Create the covariance matrix K
K <- matrix(0, nrow = 10, ncol = 10)
for (i in 1:10) {
for (j in 1:10) {
K[i,j] <- median(post$etasq) * exp(-median(post$rhosq) * islandsDistMatrix[i,j]^2)
}
}
diag(K) <- median(post$etasq) + 0.01
# 2. Convert K to a correlation matrix.
Rho <- round(cov2cor(K), 2)
# Add row and column names for convience.
colnames(Rho) <- c("Ml", "Ti", "SC", "Ya", "Fi", "Tr", "Ch", "Mn", "To", "Ha")
rownames(Rho) <- colnames(Rho)
Rho
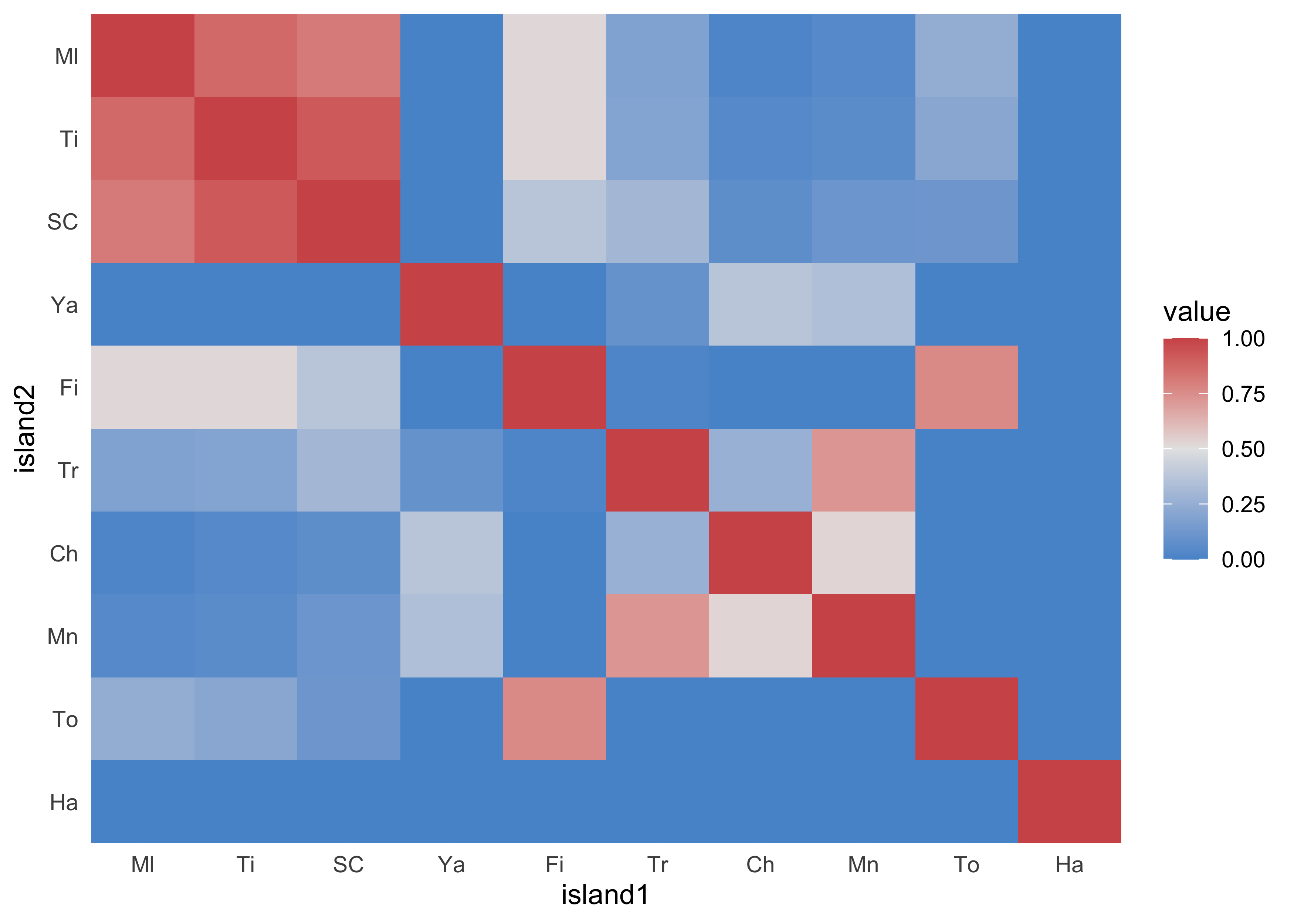
#> Ml Ti SC Ya Fi Tr Ch Mn To Ha
#> Ml 1.00 0.87 0.81 0.00 0.52 0.18 0.02 0.04 0.24 0
#> Ti 0.87 1.00 0.92 0.00 0.52 0.19 0.04 0.06 0.21 0
#> SC 0.81 0.92 1.00 0.00 0.37 0.30 0.07 0.11 0.12 0
#> Ya 0.00 0.00 0.00 1.00 0.00 0.09 0.37 0.34 0.00 0
#> Fi 0.52 0.52 0.37 0.00 1.00 0.02 0.00 0.00 0.76 0
#> Tr 0.18 0.19 0.30 0.09 0.02 1.00 0.26 0.72 0.00 0
#> Ch 0.02 0.04 0.07 0.37 0.00 0.26 1.00 0.53 0.00 0
#> Mn 0.04 0.06 0.11 0.34 0.00 0.72 0.53 1.00 0.00 0
#> To 0.24 0.21 0.12 0.00 0.76 0.00 0.00 0.00 1.00 0
#> Ha 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 1
- a cluster of highly correlative islands Ml, Ti, and SC
long_Rho <- Rho %>%
as.data.frame() %>%
rownames_to_column(var = "island1") %>%
as_tibble() %>%
mutate(island1 = fct_inorder(island1)) %>%
pivot_longer(-island1, names_to = "island2") %>%
mutate(island2 = factor(island2, levels = levels(island1)),
island2 = fct_rev(island2))
long_Rho %>%
ggplot(aes(island1, island2)) +
geom_tile(aes(fill = value)) +
scale_fill_gradient2(low = blue, mid = "grey90", high = red, midpoint = 0.5) +
scale_x_discrete(expand = c(0, 0)) +
scale_y_discrete(expand = c(0, 0))
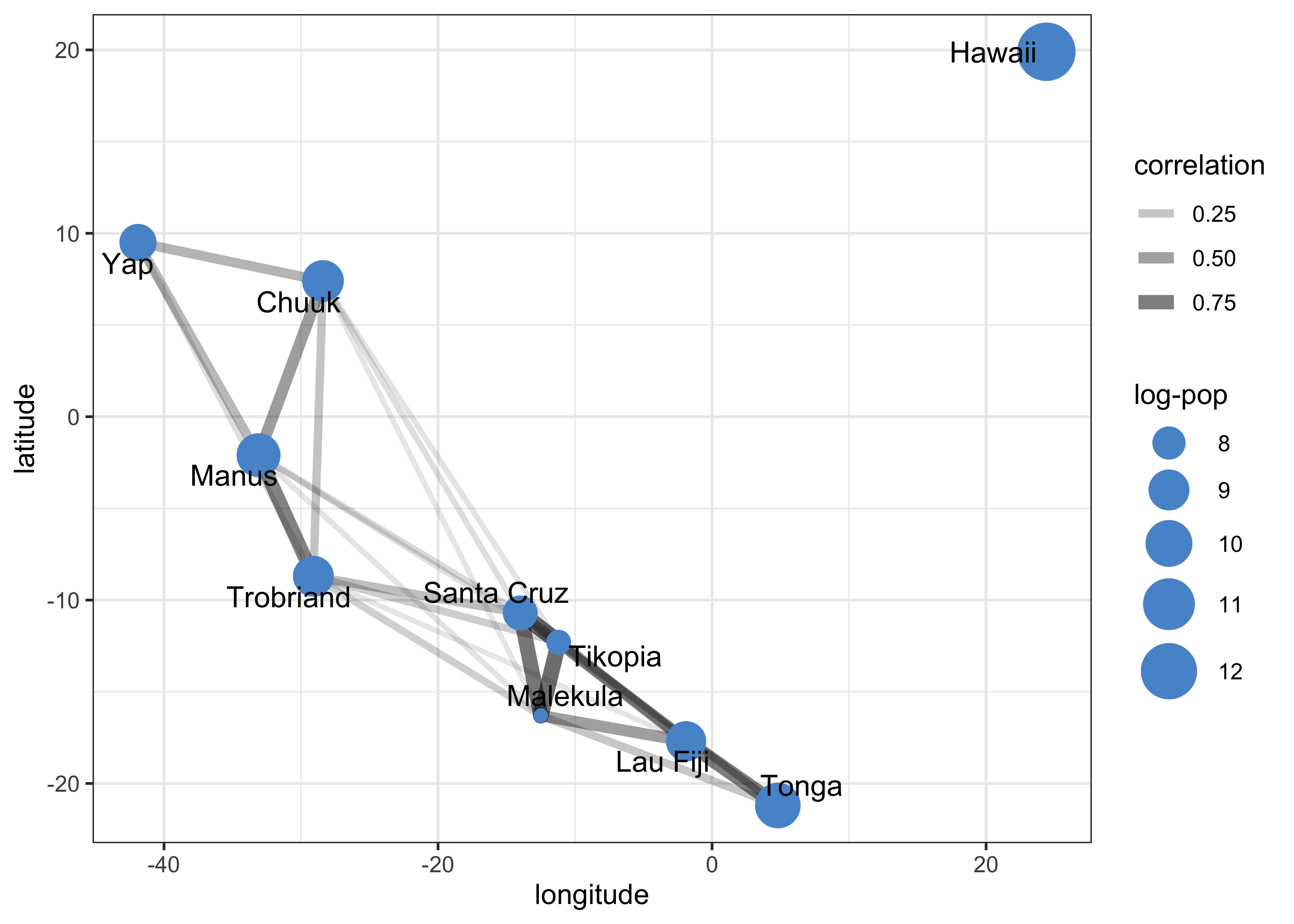
Rho_gr <- as_tbl_graph(Rho, directed = FALSE) %N>%
left_join(d %>% mutate(name = colnames(Rho)),
by = "name")
gr_layout <- create_layout(Rho_gr, layout = "nicely")
gr_layout$x <- d$lon2[match(colnames(Rho), gr_layout$name)]
gr_layout$y <- d$lat[match(colnames(Rho), gr_layout$name)]
ggraph(gr_layout) +
geom_edge_link(aes(alpha = weight, width = weight),
lineend = "round", linejoin = "round") +
geom_node_point(aes(size = logpop), color = blue) +
geom_node_text(aes(label = culture), repel = TRUE, size = 4) +
scale_edge_width_continuous(range = c(1, 3)) +
scale_edge_alpha_continuous(range = c(0.1, 0.6)) +
scale_size_continuous(range = c(2, 10)) +
theme_bw() +
labs(x = "longitude",
y = "latitude",
size = "log-pop",
edge_width = "correlation",
edge_alpha = "correlation")
logpop_seq <- seq(6, 14, length.out = 50)
lambda <- map(logpop_seq, ~ exp(post$a + post$b*.x))
lambda_median <- map_dbl(lambda, median)
lambda_pi80 <- map(lambda, PI, prob = 0.80) %>%
map_dfr(pi_to_df)
post_pred <- tibble(logpop = logpop_seq,
lambda_median = lambda_median) %>%
bind_cols(lambda_pi80) %>%
mutate(x10_percent = map_dbl(x10_percent, ~ max(c(.x, 10))),
x90_percent = map_dbl(x90_percent, ~ min(c(.x, 75))))
gr_layout <- create_layout(Rho_gr, layout = "nicely")
gr_layout$x <- d$logpop[match(colnames(Rho), gr_layout$name)]
gr_layout$y <- d$total_tools[match(colnames(Rho), gr_layout$name)]
ggraph(gr_layout) +
geom_ribbon((aes(x = logpop, ymin = x10_percent, ymax = x90_percent)),
data = post_pred,
alpha = 0.1) +
geom_line(aes(x = logpop, y = lambda_median),
data = post_pred,
color = grey, size = 1, alpha = 0.7, lty = 2) +
geom_edge_link(aes(alpha = weight, width = weight),
lineend = "round", linejoin = "round") +
geom_node_point(aes(size = logpop), color = blue) +
geom_node_text(aes(label = culture), repel = TRUE, size = 4) +
scale_edge_width_continuous(range = c(1, 3)) +
scale_edge_alpha_continuous(range = c(0.1, 0.6)) +
scale_size_continuous(range = c(2, 10)) +
scale_x_continuous(limits = c(6, 13), expand = c(0, 0)) +
scale_y_continuous(limits = c(10, 75), expand = c(0, 0)) +
theme_bw() +
labs(x = "log population",
y = "total tools",
size = "log-pop",
edge_width = "correlation",
edge_alpha = "correlation",
title = "Association of log-population and total tools, accouting for spatial correlations",
subtitle = "The dashed line is the median of the posterior prediction of the total tools given the log-population\n(ignoring the spatial variance), surrounded by the 80 PI.")
#> Warning: Removed 11 row(s) containing missing values (geom_path).
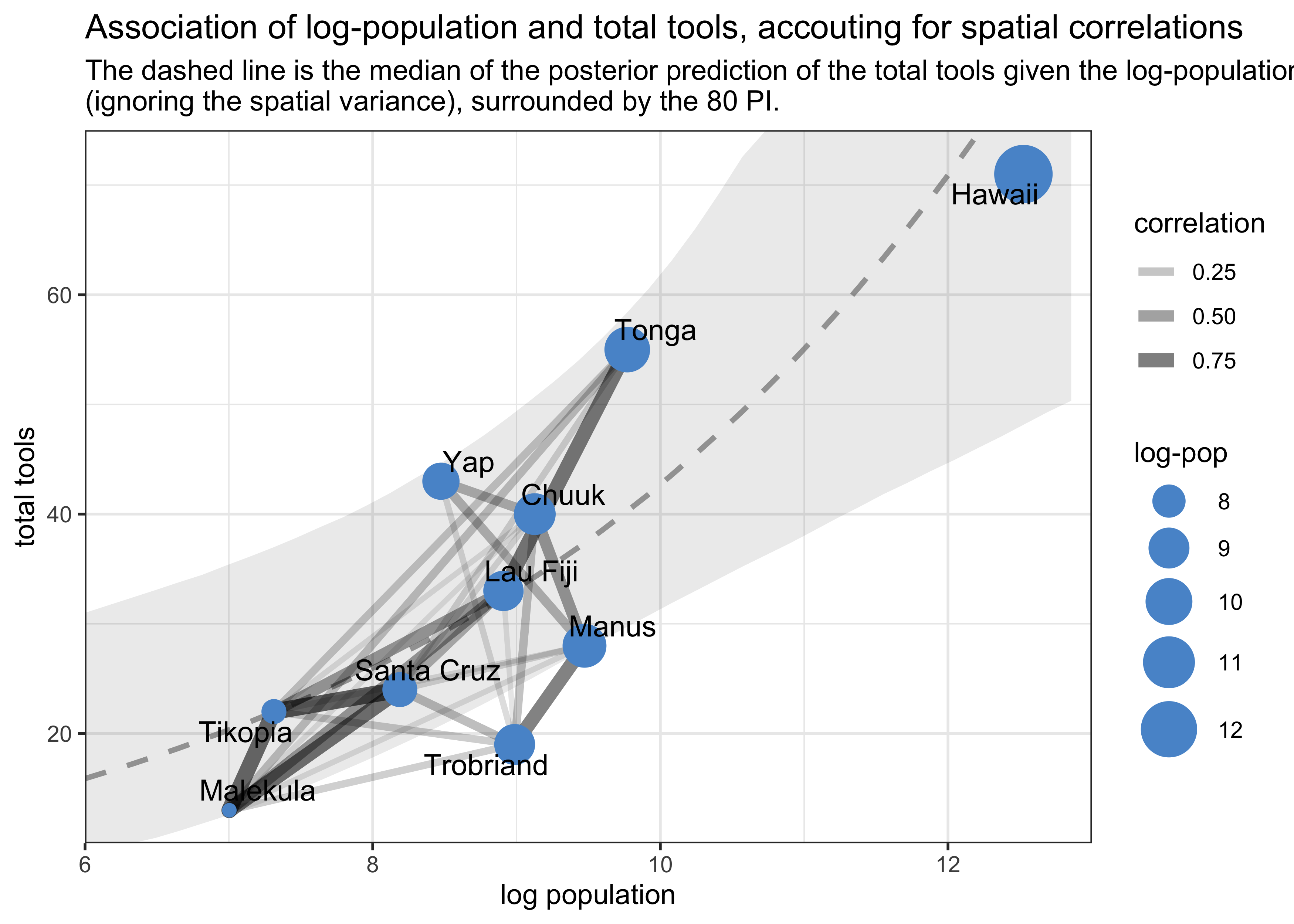
- interpretation of above plots:
- in the first, we see that Ml, Ti, and SC are all very close together spatially and have a high correlation of their varying intercepts
- the second shows that these three cultures are below the
expected number of tools per their population
- they they all lie below the expectation and are so close together is consistent with spatial covariance
13.4.2 Other kinds of “distance”
- other examples of a continuous variable for the varying effect:
- phyolgenetic distance
- network distance
- cyclic covariation of time
- build the covariance matrix with a periodic function such as sine or cosine
- also possible to have more than one dimension of distance in the
same model
- gets merged into a single covariance matrix