Chapter 12. Multilevel Models
- multi-level models remember features of each cluster in the data as
they learn about all of the clusters
- depending on the variation across clusters, the model pools information across clusters
- the pooling improves estimates about each cluster
- benefits of the multilevel approach:
- improved estimates for repeat sampling
- improved estimates for imbalance in sampling
- estimates of variation
- avoid averaging and retain variation
- multilevel regression should be the default approach
- this chapter starts with the foundations and the following two are more advanced types of multilevel models
12.1 Example: Multilivel tadpoles
- example: Reed frog tadpole mortality
surv: number or survivorscount: initial number
data("reedfrogs")
d <- as_tibble(reedfrogs)
skimr::skim(d)
| Name | d |
| Number of rows | 48 |
| Number of columns | 5 |
| _______________________ | |
| Column type frequency: | |
| factor | 2 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Data summary
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| pred | 0 | 1 | FALSE | 2 | no: 24, pre: 24 |
| size | 0 | 1 | FALSE | 2 | big: 24, sma: 24 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| density | 0 | 1 | 23.33 | 10.38 | 10.00 | 10.0 | 25.00 | 35.00 | 35 | ▇▁▇▁▇ |
| surv | 0 | 1 | 16.31 | 9.88 | 4.00 | 9.0 | 12.50 | 23.00 | 35 | ▇▂▂▂▃ |
| propsurv | 0 | 1 | 0.72 | 0.27 | 0.11 | 0.5 | 0.89 | 0.92 | 1 | ▁▂▂▁▇ |
- there is a lot of variation in the data
- some from experimental treatment, other sources do exist
- each row is a fish tank that is the experimental environment
- each tank is a cluster variable and there are repeated measures from each
- each tank may have a different baseline level of survival, but
don’t want to treat them as completely unrelated
- a dummy variable for each tank would be the wrong solution
- varying intercepts model: a multilevel model that estimates an
intercept for each tank and the variation among tanks
- for each cluster in the data, use a unique intercept parameter, adaptively learning the prior common to all of the intercepts
- what is learned about each cluster informs all the other clusters
- model for predicting tadpole mortality in each tank (nothing new)
$$ s_i \sim \text{Binomial}(n_i, p_i) $$ $$ \text{logit}(p_i) = \alpha_{\text{tank}[i]} $$ $$ \alpha_{\text{tank}} \sim \text{Normal}(0, 5) $$ $$ $$
d$tank <- 1:nrow(d)
stash("m12_1", {
m12_1 <- map2stan(
alist(
surv ~ dbinom(density, p),
logit(p) <- a_tank[tank],
a_tank[tank] ~ dnorm(0, 5)
),
data = d
)
})
#> Loading stashed object.
print(m12_1)
#> map2stan model
#> 1000 samples from 1 chain
#>
#> Sampling durations (seconds):
#> warmup sample total
#> chain:1 0.4 0.35 0.75
#>
#> Formula:
#> surv ~ dbinom(density, p)
#> logit(p) <- a_tank[tank]
#> a_tank[tank] ~ dnorm(0, 5)
#>
#> WAIC (SE): 1023 (42.9)
#> pWAIC: 49.38
precis(m12_1, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_tank[1] 2.507409171 1.1524623 0.9310192 4.3793634 1169.7322 1.0026114
#> a_tank[2] 5.612838269 2.7132323 2.1955474 10.5489376 899.3637 1.0008726
#> a_tank[3] 0.955606971 0.7242290 -0.1401785 2.1775927 1460.8578 0.9990305
#> a_tank[4] 5.679923948 2.8455974 2.1598744 10.9713731 699.5095 1.0000048
#> a_tank[5] 2.499838112 1.1727475 0.7699405 4.5863504 1188.1499 1.0000598
#> a_tank[6] 2.517113412 1.1294493 0.9249108 4.4658574 1464.1416 0.9992449
#> a_tank[7] 5.901717876 2.8211771 2.1485777 10.9912848 806.4153 1.0008287
#> a_tank[8] 2.524943494 1.1865708 0.9016065 4.6178622 1326.8743 0.9992859
#> a_tank[9] -0.434933639 0.6992316 -1.6037425 0.6736126 1861.5726 0.9990238
#> a_tank[10] 2.553526682 1.2725275 0.9580848 4.7399367 811.3452 1.0006649
#> a_tank[11] 0.928007321 0.7012304 -0.1163728 2.0012056 1709.8194 0.9990331
#> a_tank[12] 0.429178891 0.6465231 -0.5735093 1.4651116 1741.1834 0.9991630
#> a_tank[13] 0.920048116 0.7780154 -0.2456616 2.2674043 2287.5157 0.9989996
#> a_tank[14] -0.004138478 0.6422798 -1.0314039 0.9954486 2099.6606 0.9996044
#> a_tank[15] 2.534309631 1.1870475 0.9453520 4.6536340 1017.3940 0.9990010
#> a_tank[16] 2.562492002 1.1979390 0.9383206 4.6150822 904.8553 0.9993926
#> [ reached 'max' / getOption("max.print") -- omitted 32 rows ]
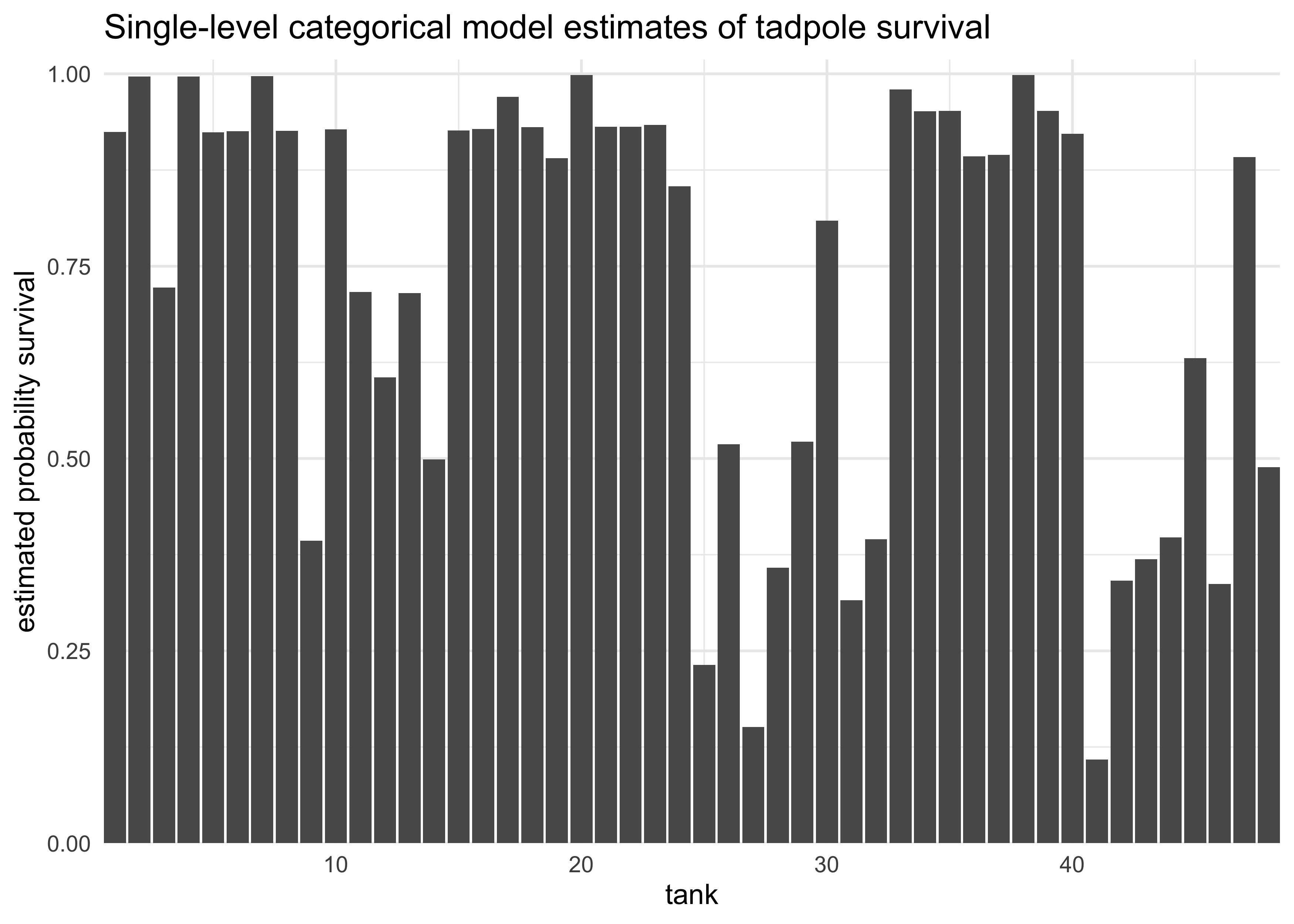
- can get expected mortality for each tank by taking the logistic of the coefficients
logistic(coef(m12_1)) %>%
enframe() %>%
mutate(name = str_remove_all(name, "a_tank\$$|\$$"),
name = as.numeric(name)) %>%
ggplot(aes(x = name, y = value)) +
geom_col() +
scale_x_continuous(expand = c(0, 0)) +
scale_y_continuous(expand = expansion(mult = c(0, 0.02))) +
labs(x = "tank",
y = "estimated probability survival",
title = "Single-level categorical model estimates of tadpole survival")
- fit a multilevel model by adding a prior for the
a_tankparameters as a function of its own parameters- now the priors have prior distributions, creating two levels of priors
$$ s_i \sim \text{Binomial}(n_i, p_i) $$ $$ \text{logit}(p_i) = \alpha_{\text{tank}[i]} $$ $$ \alpha_{\text{tank}} \sim \text{Normal}(\alpha, \sigma) $$ $$ \alpha \sim \text{Normal}(0, 1) $$ $$ \sigma \sim \text{HalfCauchy}(0, 1) $$
stash("m12_2", {
m12_2 <- map2stan(
alist(
surv ~ dbinom(density, p),
logit(p) <- a_tank[tank],
a_tank[tank] ~ dnorm(a, sigma),
a ~ dnorm(0, 1),
sigma ~ dcauchy(0, 1)
),
data = d,
iter = 4000,
chains = 4,
cores = 1
)
})
#> Loading stashed object.
print(m12_2)
#> map2stan model
#> 8000 samples from 4 chains
#>
#> Sampling durations (seconds):
#> warmup sample total
#> chain:1 0.49 0.57 1.06
#> chain:2 0.96 0.80 1.77
#> chain:3 0.81 0.78 1.59
#> chain:4 0.86 0.79 1.65
#>
#> Formula:
#> surv ~ dbinom(density, p)
#> logit(p) <- a_tank[tank]
#> a_tank[tank] ~ dnorm(a, sigma)
#> a ~ dnorm(0, 1)
#> sigma ~ dcauchy(0, 1)
#>
#> WAIC (SE): 1010 (37.9)
#> pWAIC: 37.83
precis(m12_2, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_tank[1] 2.1157682 0.8639729 0.85470075 3.5960058 12139.08 0.9998409
#> a_tank[2] 3.0669080 1.1168484 1.47355820 4.9864858 10998.05 0.9995613
#> a_tank[3] 0.9913351 0.6786515 -0.05967850 2.1209062 15763.42 0.9997261
#> a_tank[4] 3.0461374 1.1196130 1.42470945 4.9792648 11298.69 0.9997267
#> a_tank[5] 2.1296066 0.8687028 0.84447299 3.6169937 14282.14 0.9997553
#> a_tank[6] 2.1356804 0.8839724 0.84089789 3.6140100 13062.73 0.9997334
#> a_tank[7] 3.0432717 1.1007268 1.47339067 4.9349950 11197.91 0.9995695
#> a_tank[8] 2.1174578 0.8736278 0.84567072 3.6043714 13690.49 0.9996902
#> a_tank[9] -0.1801081 0.6016949 -1.15426295 0.7651194 17013.37 0.9996434
#> a_tank[10] 2.1222757 0.8745242 0.83836130 3.6369076 11732.00 0.9998723
#> a_tank[11] 1.0005859 0.6720581 -0.04092572 2.0949262 16403.51 0.9999348
#> a_tank[12] 0.5737242 0.6156076 -0.39848147 1.5714115 17386.07 1.0000767
#> a_tank[13] 0.9925635 0.6643335 -0.03262575 2.0959440 14427.52 0.9996798
#> a_tank[14] 0.1937834 0.6183100 -0.78266182 1.1916493 17018.95 0.9997141
#> a_tank[15] 2.1205258 0.8764071 0.82664938 3.5924005 13884.02 0.9996914
#> a_tank[16] 2.1278411 0.8625659 0.85117774 3.6198355 13911.59 0.9997513
#> [ reached 'max' / getOption("max.print") -- omitted 34 rows ]
- interpretation:
- $\alpha$: one overall sample intercept
- $\sigma$: variance among tanks
- 48 per-tank intercepts
compare(m12_1, m12_2)
#> WAIC SE dWAIC dSE pWAIC weight
#> m12_2 1009.876 37.94391 0.000 NA 37.83139 0.998797924
#> m12_1 1023.321 42.90494 13.445 6.642773 49.38454 0.001202076
- from the comparison, see that the multilevel model only has ~38
effective parameters
- 12 fewer than the single-level model because the prior assigned
to each intercept shrinks them all towards the mean $\alpha$
- $\alpha$ is acting like a regularizing prior, but it has been learned from the data
- 12 fewer than the single-level model because the prior assigned
to each intercept shrinks them all towards the mean $\alpha$
- plot and compare the posterior medians from both models
post <- extract.samples(m12_2)
d %>%
mutate(propsurv_estimate = logistic(apply(post$a_tank, 2, median)),
pop_size = case_when(
density == 10 ~ "small tank",
density == 25 ~ "medium tank",
density == 35 ~ "large tank"
),
pop_size = fct_reorder(pop_size, density)) %>%
ggplot(aes(tank)) +
facet_wrap(~ pop_size, nrow = 1, scales = "free_x") +
geom_hline(yintercept = logistic(median(post$a)),
lty = 2, color = dark_grey) +
geom_linerange(aes(x = tank, ymin = propsurv, ymax = propsurv_estimate),
color = light_grey, size = 1) +
geom_point(aes(y = propsurv),
color = grey, size = 1) +
geom_point(aes(y = propsurv_estimate),
color = purple, size = 1) +
labs(x = "tank",
y = "proportion surivival",
title = "Propotion of survival among tadpoles from different tanks.")
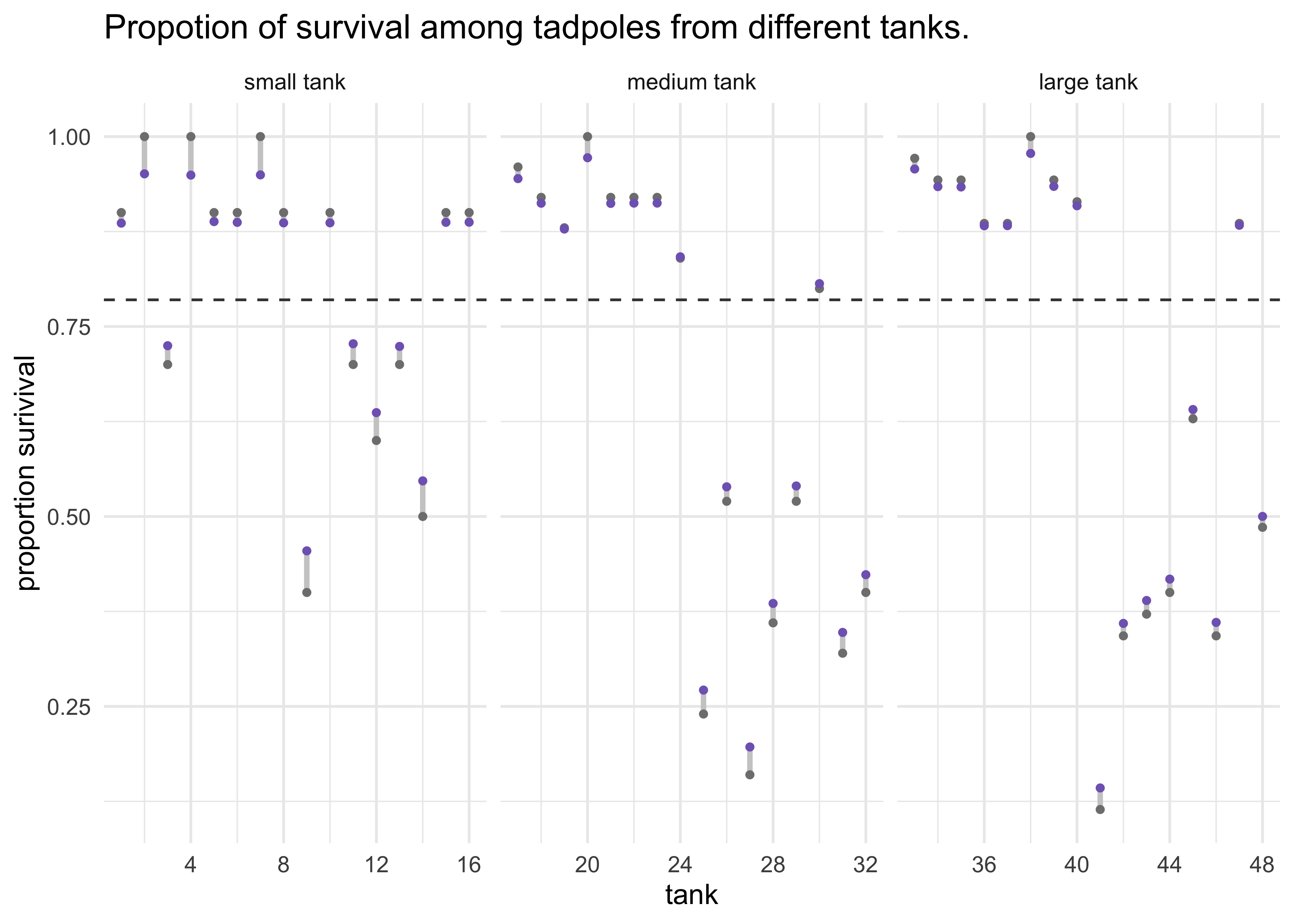
- comments on above plot:
- note that all of the purple points $\alpha_\text{tank}$ are
skewed towards to the dashed line $\alpha$
- this is often called shrinkage and comes from regularization
- note that the smaller tanks have shifted more than in the larger
tanks
- there are fewer starting tadpoles, so the shrinkage has a stronger effect
- the shift of the purple points is large the further the empirical value (grey points) are from the dashed line $\alpha$
- note that all of the purple points $\alpha_\text{tank}$ are
skewed towards to the dashed line $\alpha$
- sample from the posterior distributions:
- first plot 100 Gaussian distributions from samples of the posteriors for $\alpha$ and $\sigma$
- then sample 8000 new log-odds of survival for individual tanks
x <- seq(-3, 5, length.out = 300)
log_odds_gaussian_samples <- map_dfr(1:100, function(i) {
tibble(i, x, prob = dnorm(x, post$a[i], post$sigma[i]))
})
p1 <- log_odds_gaussian_samples %>%
ggplot(aes(x, prob, group = factor(i))) +
geom_line(alpha = 0.5, size = 0.1) +
labs(x = "log-odds survival",
y = "density",
title = "Sampled probability density curves")
p2 <- tibble(sim_tanks = logistic(rnorm(8000, post$a, post$sigma))) %>%
ggplot(aes(sim_tanks)) +
geom_density(size = 1, fill = grey, alpha = 0.5) +
scale_x_continuous(expand = c(0, 0)) +
scale_y_continuous(expand = expansion(mult = c(0, 0.02))) +
labs(x = "probability survive",
y = "density",
title = "Simulated survival proportions")
p1 | p2
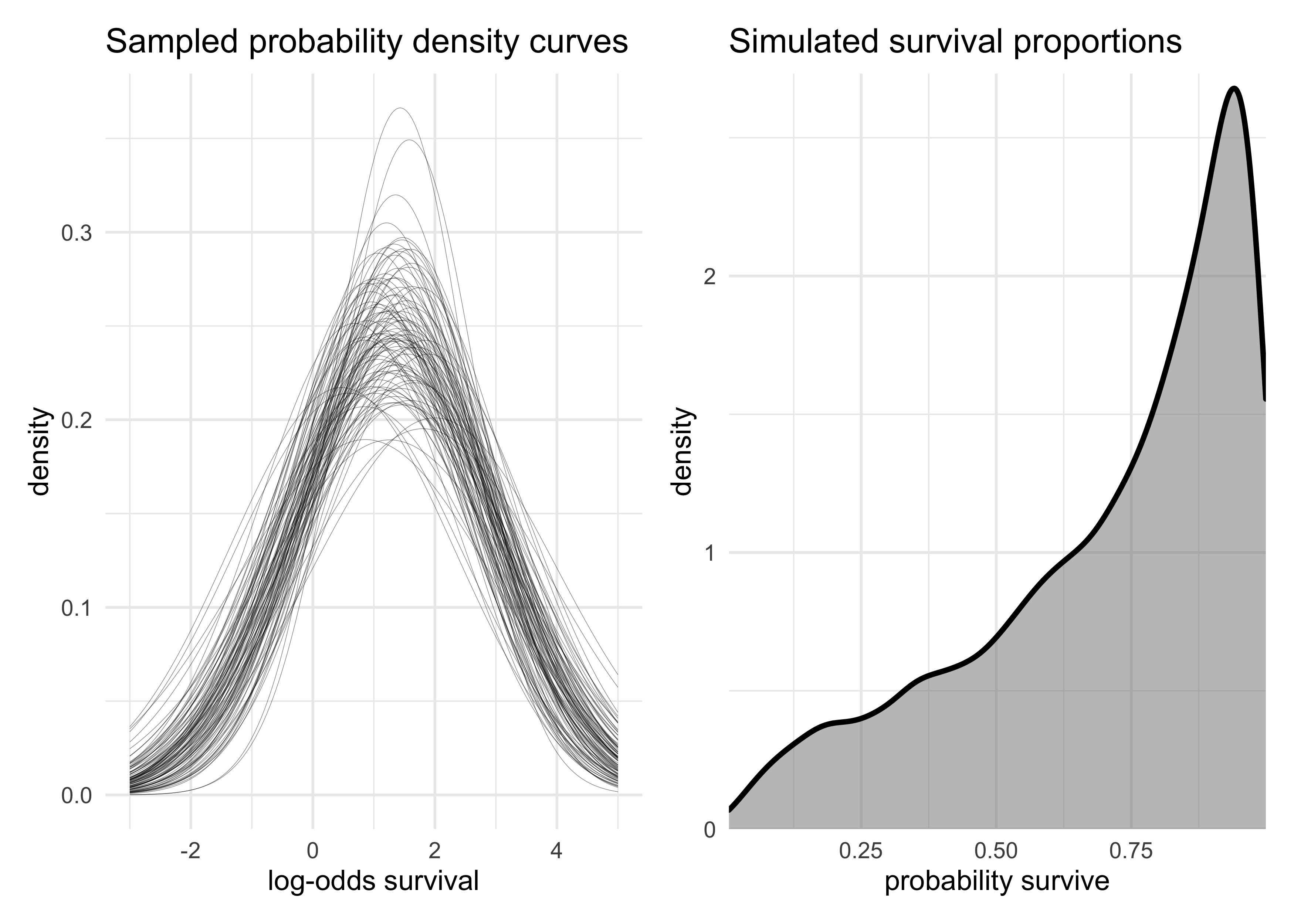
- there is uncertainty about both the location $\alpha$ and scale
$\sigma$ of the population distribution of log-odds of survival
- this uncertainty is propagated into the simulated probabilities of survival
12.2 Varying effects and the underfitting/overfitting trade-off
- “Varying intercepts are just regularized estimates, but adaptivelyy
regulraized by estimating how diverse the cluster are while
estimating the features of each cluster.”
- varying effect estimates are more accurate estimates of the individual cluster intercepts
- partial pooling helps prevent overfitting and underfitting
- pooling all of the tanks into a single intercept would make an underfit model
- having completely separate intercepts for each tank would overfit
- demonstration: simulate tadpole data so we know the true per-pond
survival probabilities
- this is also a demonstration of the important skill of simulation and model validation
12.2.1 The model
- we will use the same multilevel binomial model as before (using “ponds” instead of “tanks”)
$$ s_i \sim \text{Binomial}(n_i, p_i) $$ $$ \text{logit}(p_i) = \alpha_{\text{pond[i]}} $$ $$ \alpha_\text{pond} \sim \text{Normal}(\alpha, \sigma) $$ $$ \alpha \sim \text{Normal}(0, 1) $$ $$ \sigma \sim \text{HalfCauchy}(0, 1) $$ - need to assign values for: * $\alpha$: the average log-odds of survival for all of the ponds * $\sigma$: the standard deviation of the distribution of log-odds of survival among ponds * $\alpha_\text{pond}$: the individual pond intercepts * $n_i$: the number of tadpoles per pond
12.2.2 Assign values to the parameters
- steps in code:
- initialize $\alpha$, $\sigma$, number of ponds, number of tadpoles per ponds
- use these parameters to generate $\alpha_\text{pond}$
- put data into a data frame
set.seed(0)
# 1. Initialize top level parameters.
a <- 1.4
sigma <- 1.5
nponds <- 60
ni <- as.integer(rep(c(5, 10, 25, 35), each = 15))
# 2. Sample second level parameters for each pond.
a_pond <- rnorm(nponds, mean = a, sd = sigma)
# 3. Organize into a data frame.
dsim <- tibble(pond = seq(1, nponds),
ni = ni,
true_a = a_pond)
dsim
#> # A tibble: 60 x 3
#> pond ni true_a
#> <int> <int> <dbl>
#> 1 1 5 3.29
#> 2 2 5 0.911
#> 3 3 5 3.39
#> 4 4 5 3.31
#> 5 5 5 2.02
#> 6 6 5 -0.910
#> 7 7 5 0.00715
#> 8 8 5 0.958
#> 9 9 5 1.39
#> 10 10 5 5.01
#> # … with 50 more rows
12.2.3 Simulate survivors
- simulate the binomial survival process
- each pond $i$ has $n_i$ potential survivors with probability of survival $p_i$
- from the model definition (using the logit link function), $p_i$ is:
$$ p_i = \frac{\exp(\alpha_i)}{1 + \exp(\alpha_i)} $$
dsim$si <- rbinom(nponds,
prob = logistic(dsim$true_a),
size = dsim$ni)
12.2.4 Compute the no-pooling estiamtes
- the estimates from not pooling information across ponds is the same
as calculating the proportion of survivors in each pond
- would get same values if used a dummy variable for each pond and weak priors
- calculate these value and keep on the probability scale
dsim$p_nopool <- dsim$si / dsim$ni
12.2.5 Compute the partial-pooling estimates
- now fit the multilevel model
stash("m12_3", {
m12_3 <- map2stan(
alist(
si ~ dbinom(ni, p),
logit(p) <- a_pond[pond],
a_pond[pond] ~ dnorm(a, sigma),
a ~ dnorm(0, 1),
sigma ~ dcauchy(0, 1)
),
data = dsim,
iter = 1e4,
warmup = 1000
)
})
#> Loading stashed object.
precis(m12_3, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_pond[1] 2.5053151 1.1125019 0.8842713 4.4213093 9633.975 0.9999010
#> a_pond[2] -0.4977377 0.8163556 -1.8456079 0.7864969 13954.284 0.9999419
#> a_pond[3] 2.4900753 1.0993925 0.8644481 4.3488823 11543.637 1.0000289
#> a_pond[4] 2.5089798 1.1186088 0.8810598 4.4225452 8439.085 0.9999777
#> a_pond[5] 2.5067354 1.1084190 0.8990796 4.4121802 8900.319 0.9999229
#> a_pond[6] 0.1340153 0.7981828 -1.1291088 1.3944146 15656.275 1.0004159
#> a_pond[7] 0.7728565 0.8203942 -0.5035563 2.1126640 13286.103 0.9998930
#> a_pond[8] 1.5282302 0.9254883 0.1451402 3.0931019 11202.035 0.9998956
#> a_pond[9] 2.5097866 1.1005938 0.8965482 4.3920475 8239.384 1.0000038
#> a_pond[10] 2.4819890 1.1084175 0.8572985 4.3915817 12718.861 0.9999360
#> a_pond[11] 2.4793137 1.0892989 0.8863951 4.2945049 9759.468 1.0000382
#> a_pond[12] 0.7757112 0.8560909 -0.5276722 2.1640762 13023.329 0.9998889
#> a_pond[13] -0.4880659 0.8266811 -1.8513618 0.8007399 13347.095 1.0002797
#> a_pond[14] 2.4953084 1.0840919 0.8978628 4.3287307 10575.209 0.9998917
#> a_pond[15] 0.7641494 0.8234787 -0.5361413 2.1129975 13176.483 0.9999043
#> a_pond[16] 0.2485980 0.6022535 -0.7145539 1.2187904 17888.678 0.9999321
#> [ reached 'max' / getOption("max.print") -- omitted 46 rows ]
- compute the predicted survival proportions
estimated_a_pond <- as.numeric(coef(m12_3)[1:nponds])
dsim$p_partpool <- logistic(estimated_a_pond)
- compute known survival proportions from the real $\alpha_\text{pond}$ values
dsim$p_true <- logistic(dsim$true_a)
- plot the results and compute error between the estimated and true varying effects
dsim %>%
transmute(nopool_error = abs(p_nopool - p_true),
partpool_error = abs(p_partpool - p_true),
pond, ni) %>%
pivot_longer(-c(pond, ni),
names_to = "model_type", values_to = "absolute_error") %>%
group_by(ni, model_type) %>%
mutate(avg_error = mean(absolute_error)) %>%
ungroup() %>%
ggplot(aes(x = pond, y = absolute_error)) +
facet_wrap(~ ni, scales = "free_x", nrow = 1) +
geom_line(aes(y = avg_error, color = model_type, group = model_type), size = 1.5, alpha = 0.7) +
geom_line(aes(group = factor(pond)), color = light_grey, size = 0.8) +
geom_point(aes(color = model_type)) +
scale_color_brewer(palette = "Dark2") +
theme(legend.position = c(0.9, 0.7)) +
labs(x = "pond number",
y = "absolute error",
color = "model type",
title = "Comparing the error between estimates from amnesiac and multilevel models")
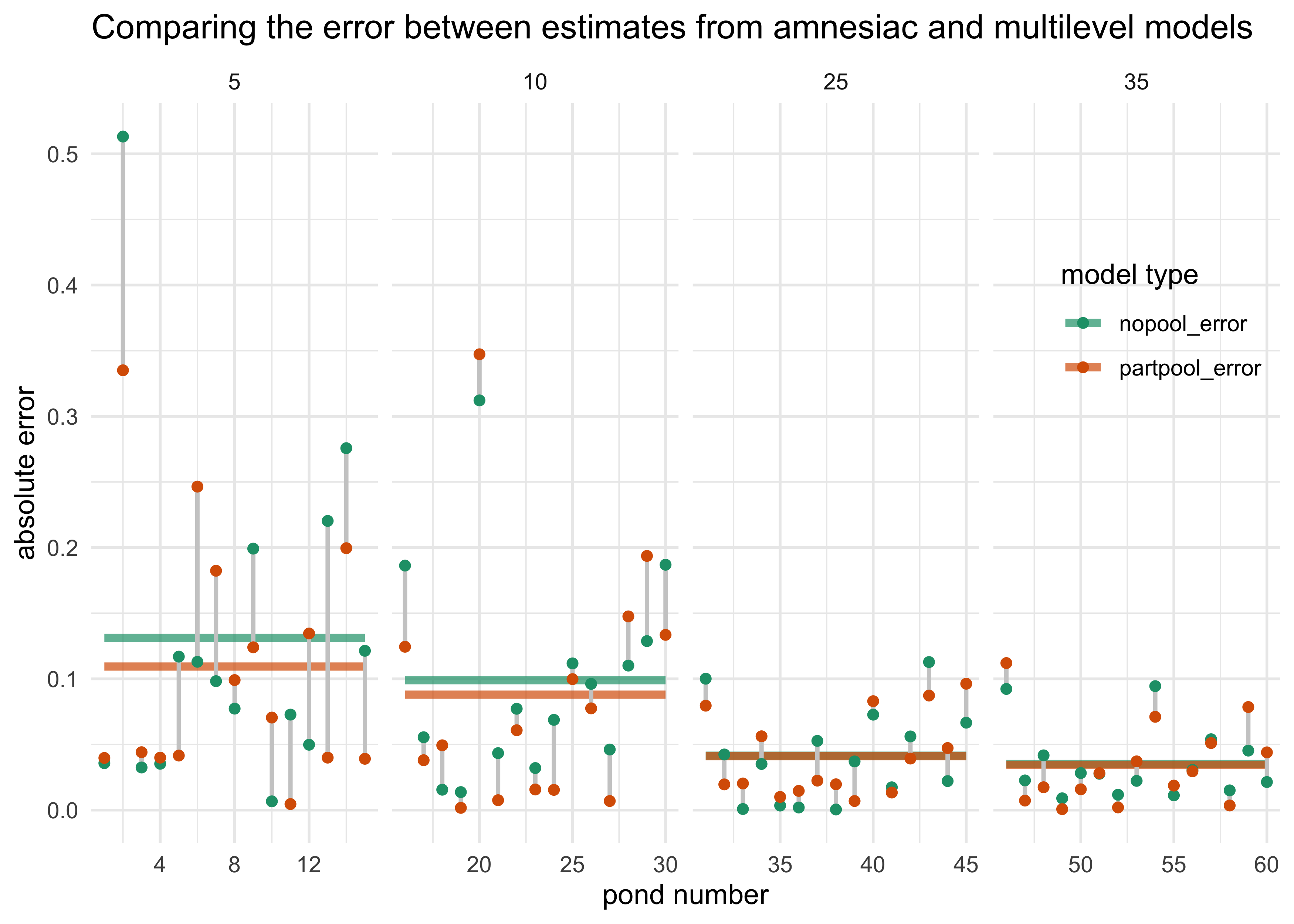
- interpretation:
- both models perform better with larger ponds becasue more data
- the partial pooling model performs better, on average, than the no pooling model
12.3 More than one type of cluster
- often are multiple clusters of data in the same model
- example: chimpanzee data
- one block for each chimp
- one block for each day of testing
12.3.1 Multilevel chimpanzees
- similar model as before
- add varying intercepts for actor
- put both the $\alpha$ and $\alpha_\text{actor}$ in the
linear model
- it is to allow for adding other varying effects
- instead of having $\alpha$ as the mean for $\alpha_\text{actor}$, the mean for $\alpha_\text{actor} = 0$ and the mean $\alpha$ is in the linear model instead
$$ L_i \sim \text{Binomial}(1, p_i) $$ $$ \text{logit}(p_i) = \alpha + \alpha_{\text{actor}[i]} + (\beta_P + \beta_{PC} C_i) P_i $$ $$ \alpha_\text{actor} \sim \text{Normal}(0, \sigma_\text{actor}) $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta_P \sim \text{Normal}(0, 10) $$ $$ \beta_{PC} \sim \text{Normal}(0, 10) $$ $$ \alpha_\text{actor} \sim \text{HalfCauchy}(0, 1) $$ $$ $$
data("chimpanzees")
d <- as_tibble(chimpanzees) %>%
select(-recipient)
stash("m12_4", {
m12_4 <- map2stan(
alist(
pulled_left ~ dbinom(1, p),
logit(p) <- a + a_actor[actor] + (bp + bpc*condition)*prosoc_left,
a_actor[actor] ~ dnorm(0, sigma_actor),
a ~ dnorm(0, 10),
bp ~ dnorm(0, 10),
bpc ~ dnorm(0, 10),
sigma_actor ~ dcauchy(0, 1)
),
data = d,
warmup = 1e3,
iter = 5e3,
chains = 4
)
})
#> Loading stashed object.
print(m12_4)
#> map2stan model
#> 16000 samples from 4 chains
#>
#> Sampling durations (seconds):
#> warmup sample total
#> chain:1 7.29 21.96 29.25
#> chain:2 7.15 23.66 30.81
#> chain:3 5.73 24.91 30.64
#> chain:4 6.07 20.77 26.85
#>
#> Formula:
#> pulled_left ~ dbinom(1, p)
#> logit(p) <- a + a_actor[actor] + (bp + bpc * condition) * prosoc_left
#> a_actor[actor] ~ dnorm(0, sigma_actor)
#> a ~ dnorm(0, 10)
#> bp ~ dnorm(0, 10)
#> bpc ~ dnorm(0, 10)
#> sigma_actor ~ dcauchy(0, 1)
#>
#> WAIC (SE): 531 (19.5)
#> pWAIC: 8.11
precis(m12_4, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_actor[1] -1.1735896 1.0012019 -2.7300473 0.23364730 2101.429 1.0012547
#> a_actor[2] 4.2071147 1.7506271 2.1320036 7.05788804 3273.263 1.0003060
#> a_actor[3] -1.4800454 1.0031659 -3.0516861 -0.05041580 2089.652 1.0013224
#> a_actor[4] -1.4757774 1.0020223 -3.0374286 -0.05192123 2099.656 1.0013993
#> a_actor[5] -1.1713944 1.0020717 -2.7413405 0.25875482 2094.726 1.0013656
#> a_actor[6] -0.2278687 0.9993039 -1.7950471 1.20604520 2102.027 1.0013138
#> a_actor[7] 1.3076865 1.0255852 -0.2800768 2.81614317 2246.178 1.0011328
#> a 0.4576892 0.9800871 -0.9178915 1.99125639 2025.506 1.0013659
#> bp 0.8231619 0.2621824 0.4136777 1.25290336 6494.962 0.9999752
#> bpc -0.1311277 0.2989366 -0.6053379 0.34261909 6701.342 1.0000421
#> sigma_actor 2.2768974 0.9825956 1.2487228 3.92728797 3201.064 1.0008673
- note that the mean population of actors $\alpha$ and the individual deviations from that mean $\alpha_\text{actor}$ must be summed to calculate the entrie intercept: $\alpha + \alpha_\text{actor}$
post <- extract.samples(m12_4)
total_a_actor <- map(1:7, ~ post$a + post$a_actor[, .x])
round(map_dbl(total_a_actor, mean), 2)
#> [1] -0.72 4.66 -1.02 -1.02 -0.71 0.23 1.77
12.3.2 Two types of cluster
- add a second cluster on
block- replicate the structure for
actor - keep only a single global mean parameter $\alpha$ and have the varying intercepts with a mean of 0
- replicate the structure for
$$ L_i \sim \text{Binomial}(1, p_i) $$ $$ \text{logit}(p_i) = \alpha + \alpha_{\text{actor}[i]} + \alpha_{\text{block}[i]} + (\beta_P + \beta_{PC} C_i) P_i $$ $$ \alpha_\text{actor} \sim \text{Normal}(0, \sigma_\text{actor}) $$ $$ \alpha_\text{block} \sim \text{Normal}(0, \sigma_\text{block}) $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta_P \sim \text{Normal}(0, 10) $$ $$ \beta_{PC} \sim \text{Normal}(0, 10) $$ $$ \alpha_\text{actor} \sim \text{HalfCauchy}(0, 1) $$ $$ \alpha_\text{block} \sim \text{HalfCauchy}(0, 1) $$ $$ $$
d$block_id <- d$block # 'block' is a reserved name in Stan.
stash("m12_5", {
m12_5 <- map2stan(
alist(
pulled_left ~ dbinom(1, p),
logit(p) <- a + a_actor[actor] + a_block[block_id] + (bp + bpc*condition)*prosoc_left,
a_actor[actor] ~ dnorm(0, sigma_actor),
a_block[block_id] ~ dnorm(0, sigma_block),
a ~ dnorm(0, 10),
bp ~ dnorm(0, 10),
bpc ~ dnorm(0, 10),
sigma_actor ~ dcauchy(0, 1),
sigma_block ~ dcauchy(0, 1)
),
data = d,
warmup = 1e3,
iter = 6e3,
chains = 4
)
})
#> Loading stashed object.
print(m12_5)
#> map2stan model
#> 20000 samples from 4 chains
#>
#> Sampling durations (seconds):
#> warmup sample total
#> chain:1 10.72 35.09 45.81
#> chain:2 7.90 29.20 37.10
#> chain:3 7.56 21.69 29.25
#> chain:4 5.48 26.69 32.18
#>
#> Formula:
#> pulled_left ~ dbinom(1, p)
#> logit(p) <- a + a_actor[actor] + a_block[block_id] + (bp + bpc *
#> condition) * prosoc_left
#> a_actor[actor] ~ dnorm(0, sigma_actor)
#> a_block[block_id] ~ dnorm(0, sigma_block)
#> a ~ dnorm(0, 10)
#> bp ~ dnorm(0, 10)
#> bpc ~ dnorm(0, 10)
#> sigma_actor ~ dcauchy(0, 1)
#> sigma_block ~ dcauchy(0, 1)
#>
#> WAIC (SE): 532 (19.7)
#> pWAIC: 10.3
precis(m12_5, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a_actor[1] -1.175064397 0.9841238 -2.7606242 0.26572500 3127.582 1.0017434
#> a_actor[2] 4.180714321 1.6729009 2.1251215 6.98239536 4813.958 1.0003135
#> a_actor[3] -1.480590647 0.9832722 -3.0791206 -0.04821499 3130.192 1.0019540
#> a_actor[4] -1.478783548 0.9836902 -3.0794615 -0.05543320 3190.286 1.0018033
#> a_actor[5] -1.174174061 0.9817486 -2.7569231 0.26783679 3081.939 1.0019227
#> a_actor[6] -0.226458711 0.9796456 -1.8242262 1.22181433 3165.682 1.0016978
#> a_actor[7] 1.316572431 1.0070672 -0.2909612 2.82251597 3279.139 1.0016276
#> a_block[1] -0.183995506 0.2313540 -0.6197247 0.07417105 4061.437 1.0011753
#> a_block[2] 0.037069316 0.1877480 -0.2391722 0.34372436 10717.249 1.0002318
#> a_block[3] 0.052974160 0.1866553 -0.2097568 0.37125753 9201.858 1.0001379
#> a_block[4] 0.004609523 0.1835454 -0.2868766 0.29340365 11224.762 1.0003635
#> a_block[5] -0.034560355 0.1870889 -0.3514144 0.23976596 10534.296 1.0004691
#> a_block[6] 0.113814284 0.1999371 -0.1362324 0.48060435 6501.544 1.0003009
#> a 0.456947755 0.9683053 -0.9553192 2.02343452 3019.549 1.0021039
#> bp 0.831444842 0.2628057 0.4108247 1.25433671 11156.767 0.9998428
#> bpc -0.141912446 0.2989153 -0.6232644 0.33594039 11467.687 1.0000441
#> [ reached 'max' / getOption("max.print") -- omitted 2 rows ]
- there was a warning message, though it can be safely ignored:
There were 11 divergent iterations during sampling. Check the chains (trace plots, n_eff, Rhat) carefully to ensure they are valid.
- interpretation:
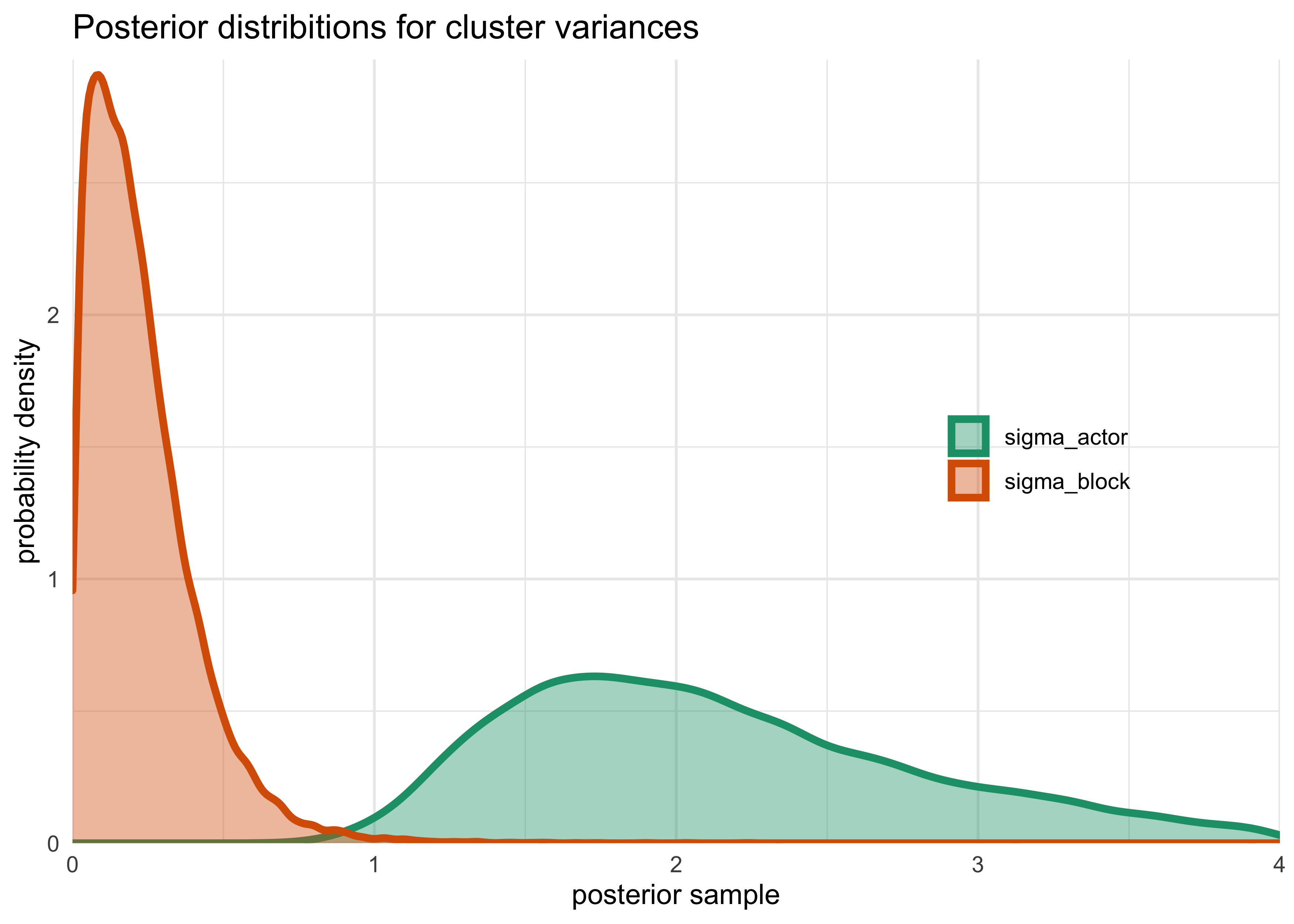
- normal to have variance of
n_effacross parameters of these more complex models - $\sigma_\text{block}$ is much smaller than
$\sigma_\text{actor}$ so there is more variation between
actors
- therefore, adding
blockhasnt added much overfitting risk
- therefore, adding
- normal to have variance of
post <- extract.samples(m12_5)
enframe(post) %>%
filter(name %in% c("sigma_actor", "sigma_block")) %>%
unnest(value) %>%
ggplot(aes(value)) +
geom_density(aes(color = name, fill = name), size = 1.4, alpha = 0.4) +
scale_x_continuous(limits = c(0, 4),
expand = c(0, 0)) +
scale_y_continuous(expand = expansion(mult = c(0, 0.02))) +
scale_color_brewer(palette = "Dark2") +
scale_fill_brewer(palette = "Dark2") +
theme(legend.title = element_blank(),
legend.position = c(0.8, 0.5)) +
labs(x = "posterior sample",
y = "probability density",
title = "Posterior distribitions for cluster variances")
#> Warning: Removed 986 rows containing non-finite values (stat_density).
compare(m12_4, m12_5)
#> WAIC SE dWAIC dSE pWAIC weight
#> m12_4 531.3511 19.50534 0.00000 NA 8.112905 0.6339337
#> m12_5 532.4494 19.66977 1.09826 1.774829 10.297114 0.3660663
- there are 7 more parameters in
m12_5thanm12_4, but thepWAIC(effective number of parameters) shows there are only about 2 more effective parameters- because the variance from
blockis so low
- because the variance from
- the models have very close WAIC values because they make very
similar predictions
blockhad very little influence on the model- keeping and reporting on both models is important to demonstrate this fact
12.3.3 Even more clusters
- MCMC can handle thousands of varying effects
- need not be shy to include a varying effect if there is theoretical
reason it would introduce variance
- overfitting risk is low as $\sigma$ for the parameters will shrink
- indicates the importance of the cluster
12.4 Multilevel posterior predictions
- model checking: a robust way to check the fit of a model is to
compare the sample to the posterior predictions
- *information criteria are also useful indicators of model flexibility and risk of overfitting
- for a multilevel model:
- should not expect to “retrodict” the sample because shrinkage will distort some predictions
- will want predictions for existing clusters of data and new clusters of data
12.4.1 Posterior prediction for same clusters
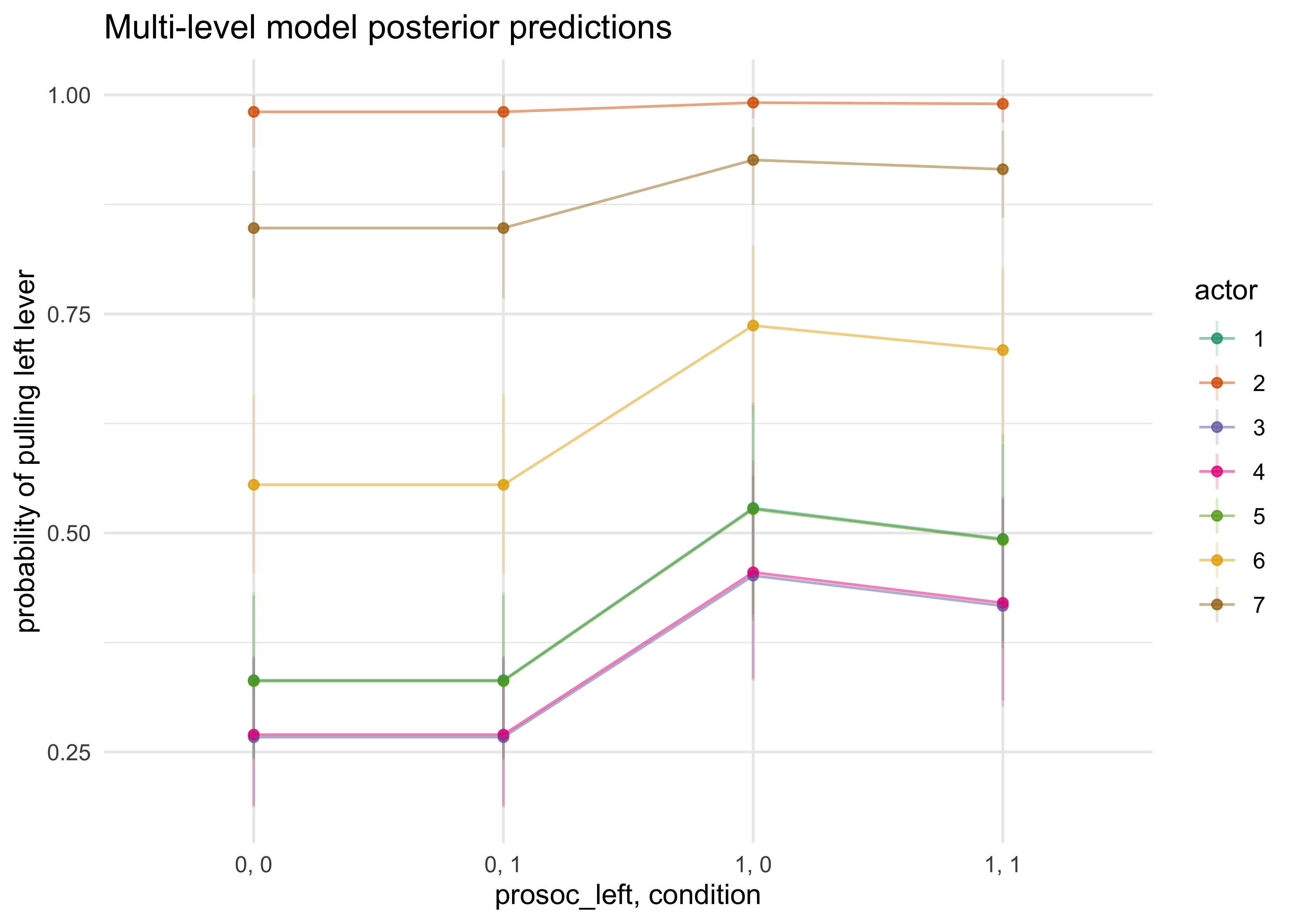
- example uing
chimpanzeesdataset and model12_4- each
actoris a cluster of the data
- each
# A data frame of all possible conditions
d_conditions <- tibble(
prosoc_left = c(0, 1, 0, 1),
condition = c(0, 0, 1, 1)
)
# A data frame of all possible conditions for each actor (chimp)
d_pred <- tibble(actor = 1:7) %>%
mutate(data = rep(list(d_conditions), 7)) %>%
unnest(data)
# make predictions
link_m12_4 <- link(m12_4, data = d_pred)
#> [ 100 / 1000 ][ 200 / 1000 ][ 300 / 1000 ][ 400 / 1000 ][ 500 / 1000 ][ 600 / 1000 ][ 700 / 1000 ][ 800 / 1000 ][ 900 / 1000 ][ 1000 / 1000 ]
d_pred %>%
mutate(post_pred_mean = apply(link_m12_4, 2, mean)) %>%
bind_cols(apply(link_m12_4, 2, PI) %>% pi_to_df()) %>%
mutate(x = paste(prosoc_left, condition, sep = ", ")) %>%
ggplot(aes(x = x, y = post_pred_mean, color = factor(actor))) +
geom_linerange(aes(ymin = x5_percent, ymax = x94_percent), alpha = 0.2) +
geom_point(alpha = 0.8) +
geom_line(aes(group = factor(actor)), alpha = 0.5) +
scale_color_brewer(palette = "Dark2") +
labs(x = "prosoc_left, condition",
y = "probability of pulling left lever",
title = "Multi-level model posterior predictions",
color = "actor")
12.4.2 Posterior prediction for new clusters
- often we do not care about the individual clusters in the data
- we don’t necessarily want predictions for the 7 chimps in the data, but for all of the species
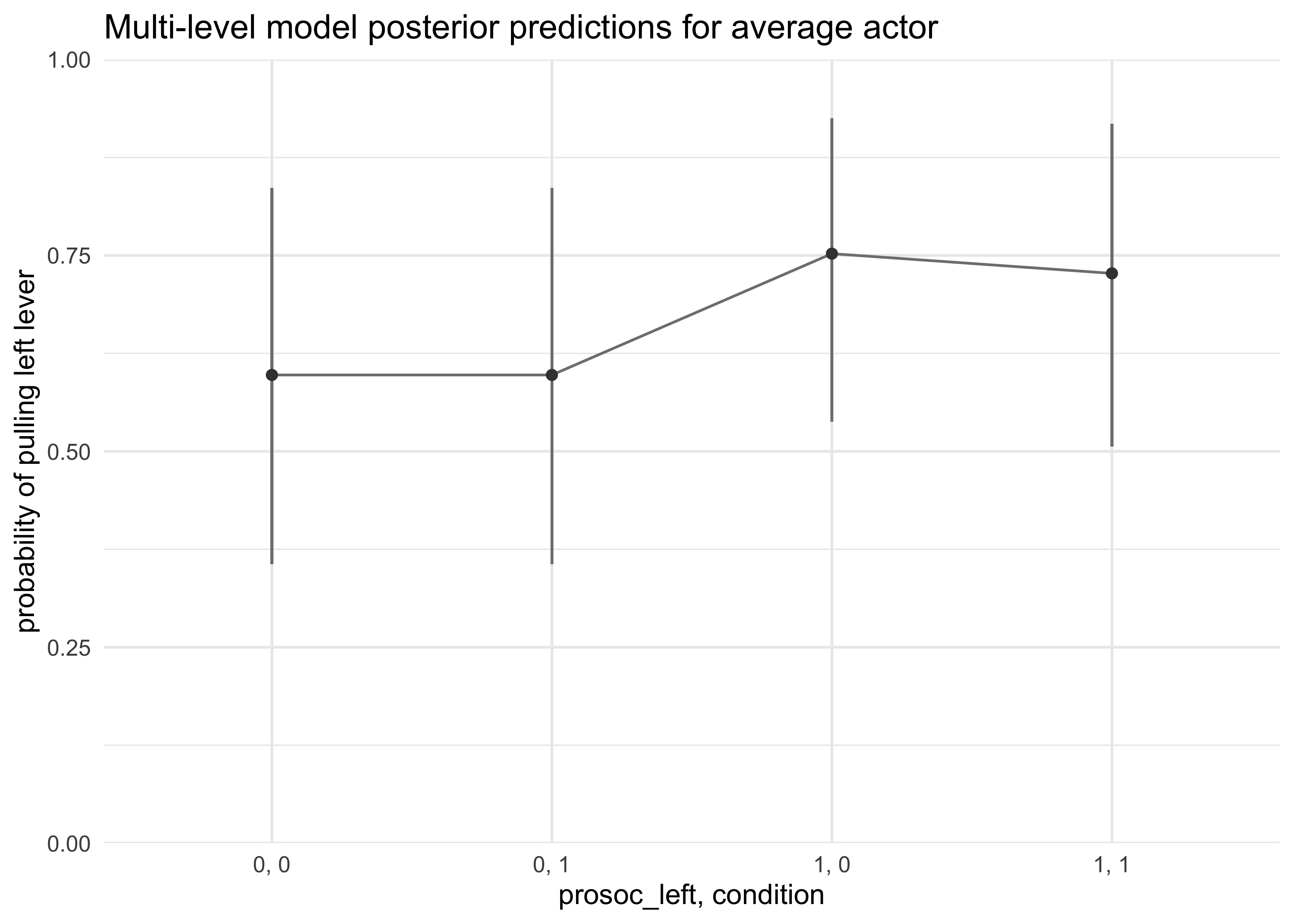
- first attempt: construct a posterior prediciton for the average
actor using $\alpha$
- however, does not show the variation among actors
d_pred$actor <- 1 # A non-zero placeholder
a_actor_zeros = matrix(0, nrow = 1e3, ncol = 7)
link_m12_4 <- link(m12_4, n = 1e3, data = d_pred,
replace = list(a_actor = a_actor_zeros))
#> [ 100 / 1000 ][ 200 / 1000 ][ 300 / 1000 ][ 400 / 1000 ][ 500 / 1000 ][ 600 / 1000 ][ 700 / 1000 ][ 800 / 1000 ][ 900 / 1000 ][ 1000 / 1000 ]
d_pred %>%
mutate(x = paste(prosoc_left, condition, sep = ", "),
pred_p_mean = apply(link_m12_4, 2, mean)) %>%
bind_cols(apply(link_m12_4, 2, PI, prob = 0.8) %>% pi_to_df()) %>%
ggplot(aes(x = x, y = pred_p_mean)) +
geom_linerange(aes(ymin = x10_percent, ymax = x90_percent), color = grey) +
geom_line(aes(group = factor(actor)), color = grey) +
geom_point(color = dark_grey) +
scale_y_continuous(limits = c(0, 1), expand = c(0, 0)) +
labs(x = "prosoc_left, condition",
y = "probability of pulling left lever",
title = "Multi-level model posterior predictions for average actor")
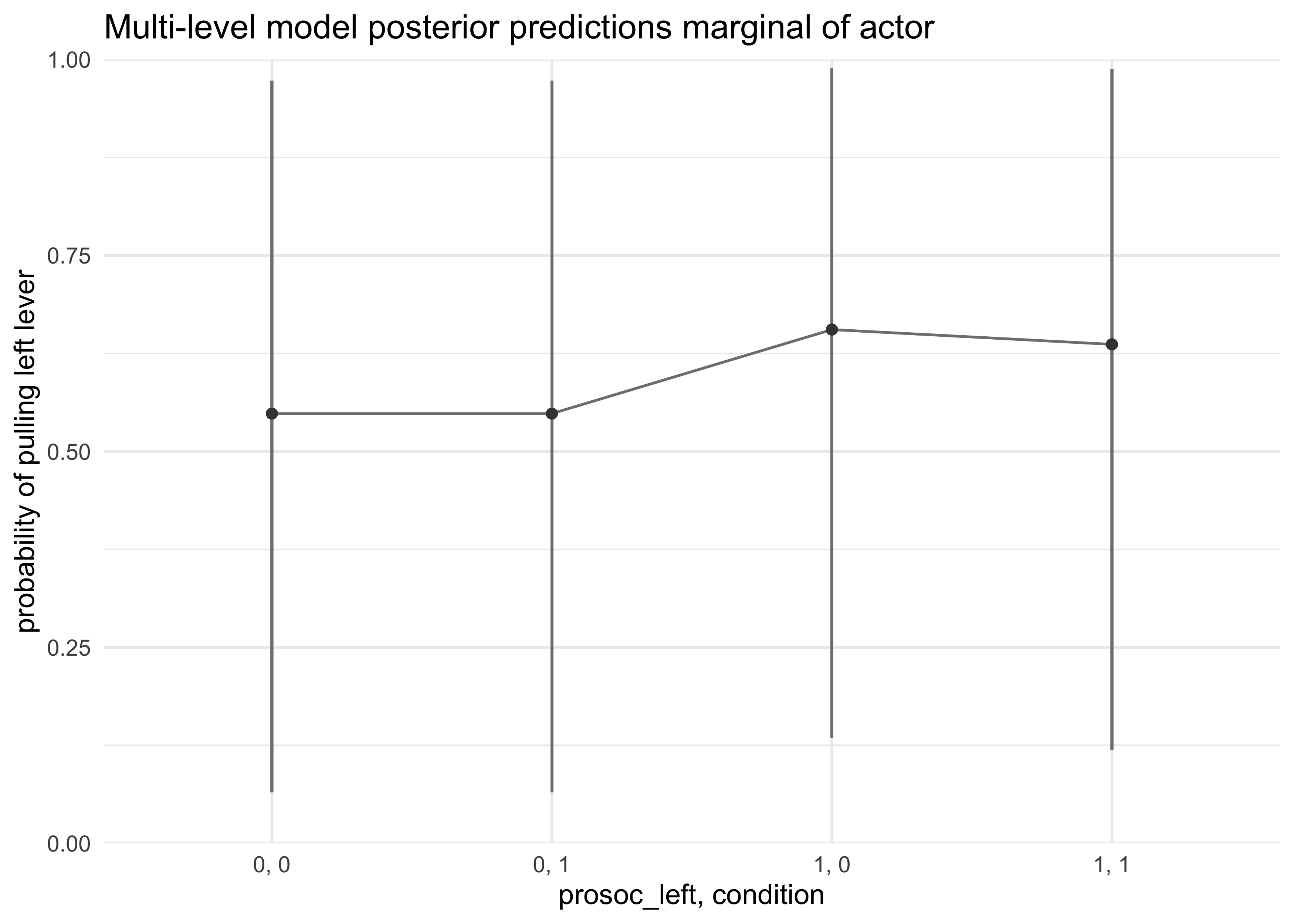
- second attempt: show variation amongst actors by including the
sigma_actorin the calculation
post <- extract.samples(m12_4)
a_actor_sims <- rnorm(7e3, 0, post$sigma_actor)
a_actor_sims <- matrix(a_actor_sims, nrow = 1e3, ncol = 7)
link_m12_4 <- link(m12_4, n = 1e3, data = d_pred,
replace = list(a_actor = a_actor_sims))
#> [ 100 / 1000 ][ 200 / 1000 ][ 300 / 1000 ][ 400 / 1000 ][ 500 / 1000 ][ 600 / 1000 ][ 700 / 1000 ][ 800 / 1000 ][ 900 / 1000 ][ 1000 / 1000 ]
d_pred %>%
mutate(x = paste(prosoc_left, condition, sep = ", "),
pred_p_mean = apply(link_m12_4, 2, mean)) %>%
bind_cols(apply(link_m12_4, 2, PI, prob = 0.8) %>% pi_to_df()) %>%
ggplot(aes(x = x, y = pred_p_mean)) +
geom_linerange(aes(ymin = x10_percent, ymax = x90_percent), color = grey) +
geom_line(aes(group = factor(actor)), color = grey) +
geom_point(color = dark_grey) +
scale_y_continuous(limits = c(0, 1), expand = c(0, 0)) +
labs(x = "prosoc_left, condition",
y = "probability of pulling left lever",
title = "Multi-level model posterior predictions marginal of actor")
- choosing which plot to use/present depends on the context and what
you are trying to learn
- the average actor plot shows the effect of treatment
- the marginal of actor plot shows how variable actors can be
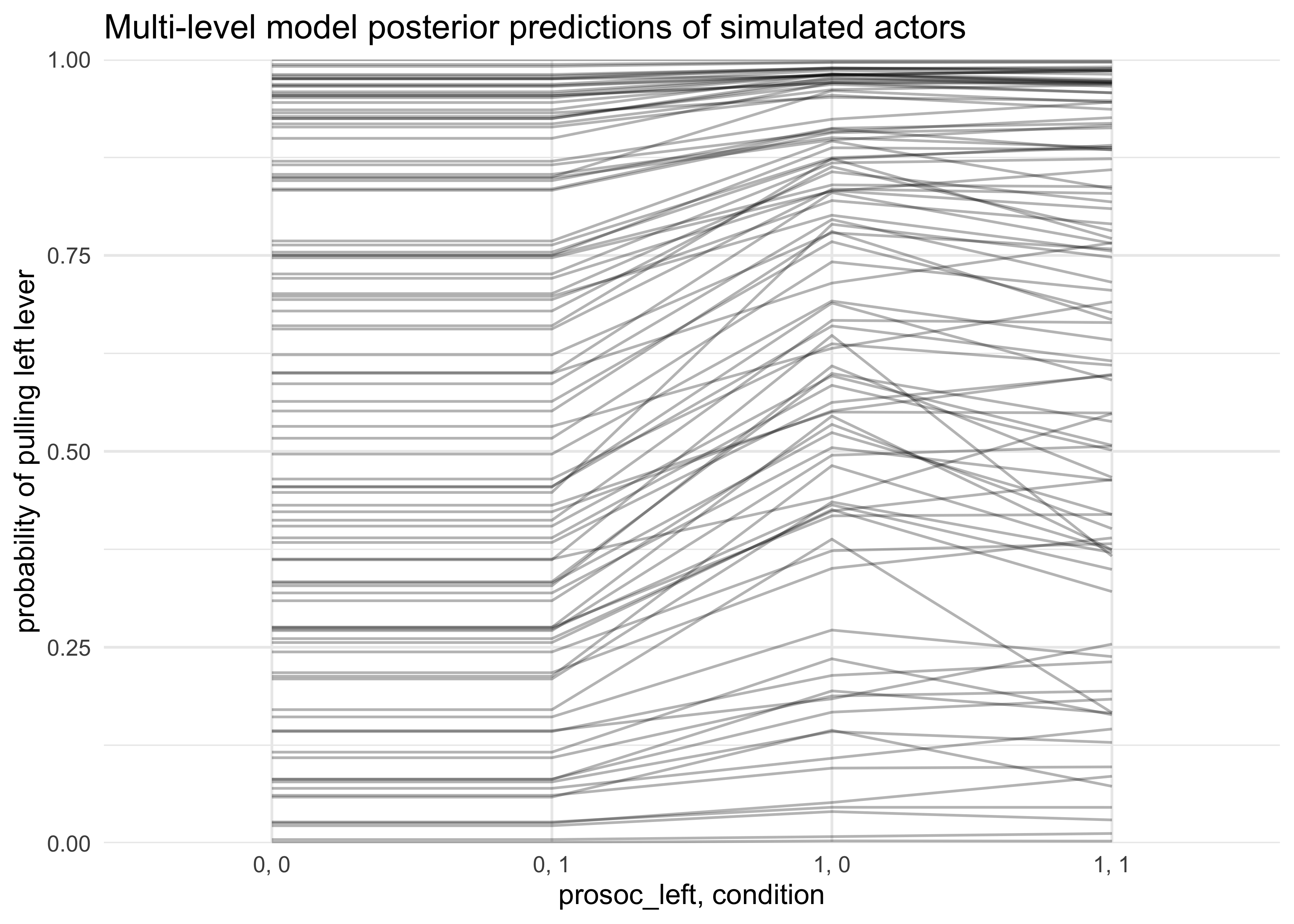
- another option is to try and show both by showing the results for a bunch of new simulated actors
post <- extract.samples(m12_4, n = 1e2)
sim_actor <- function(i) {
sim_a_actor <- rnorm(1, 0, post$sigma_actor[i])
P <- c(0, 1, 0, 1)
C <- c(0, 0, 1, 1)
p <- logistic(
post$a[i] + sim_a_actor + (post$bp[i] + post$bpc[i] * C) * P
)
return(
tibble(i = i, prosoc_left = P, condition = C, pred = p)
)
}
map_df(1:100, sim_actor) %>%
mutate(x = paste(prosoc_left, condition, sep = ", ")) %>%
ggplot(aes(x = x, y = pred)) +
geom_line(aes(group = factor(i)), alpha = 0.3) +
scale_y_continuous(limits = c(0, 1), expand = c(0, 0)) +
labs(x = "prosoc_left, condition",
y = "probability of pulling left lever",
title = "Multi-level model posterior predictions of simulated actors")
12.4.3 Focus and multilevel prediction
- can use varying effects to model over-dispersion
- example: with Oceanic societies data with an intercept for each
society
- $T$ is the
total_tools, $P$ is population, $i$ indexes each society - $\sigma_\text{society}$ is the estimate of over-dispersion among societies
- $T$ is the
- example: with Oceanic societies data with an intercept for each
society
$$ T_i \sim \text{Poisson}(\mu_i) $$ $$ \log(\mu_i) = \alpha + \alpha_{\text{society}_{[i]}} + \beta_P \log P_i $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta_P \sim \text{Normal}(0, 1) $$ $$ \alpha_\text{society} \sim \text{Normal}(0, \sigma_\text{society}) $$ $$ \sigma_\text{society} \sim \text{HalfCauchy}(0, 1) $$
data("Kline")
d <- as_tibble(Kline) %>%
janitor::clean_names() %>%
mutate(logpop = log(population),
society = row_number())
stash("m12_6", {
m12_6 <- map2stan(
alist(
total_tools ~ dpois(mu),
log(mu) <- a + a_society[society] + bp * logpop,
a ~ dnorm(0, 10),
bp ~ dnorm(0, 1),
a_society[society] ~ dnorm(0, sigma_society),
sigma_society ~ dcauchy(0, 1)
),
data = d,
iter = 4e3,
chains = 3
)
})
#> Loading stashed object.
precis(m12_6, depth = 2)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> a 1.08087255 0.72924196 -0.10532809 2.18699862 1885.393 1.001674
#> bp 0.26289831 0.07876416 0.14412084 0.39184726 1910.748 1.001932
#> a_society[1] -0.19505324 0.24046486 -0.59548503 0.15615568 2903.022 1.000199
#> a_society[2] 0.04898760 0.21650512 -0.28245864 0.39230167 2790.881 1.000004
#> a_society[3] -0.03841012 0.19526471 -0.35652854 0.26649157 3589.386 1.000526
#> a_society[4] 0.33023510 0.18975208 0.05272352 0.64920133 2464.826 1.000444
#> a_society[5] 0.04739732 0.17362694 -0.22459034 0.32530026 3321.421 1.000341
#> a_society[6] -0.31475130 0.20089950 -0.65113728 -0.01907456 3195.275 1.000715
#> a_society[7] 0.14637256 0.17070312 -0.11907974 0.42550859 3139.055 1.000605
#> a_society[8] -0.16791652 0.17754775 -0.46365739 0.10419958 3416.196 1.001071
#> a_society[9] 0.27621050 0.17103703 0.01596246 0.55890844 2629.650 1.002193
#> a_society[10] -0.09838891 0.27906914 -0.55327144 0.32098196 2118.158 1.002263
#> sigma_society 0.30690543 0.12302611 0.14853048 0.52860652 1546.725 1.003821
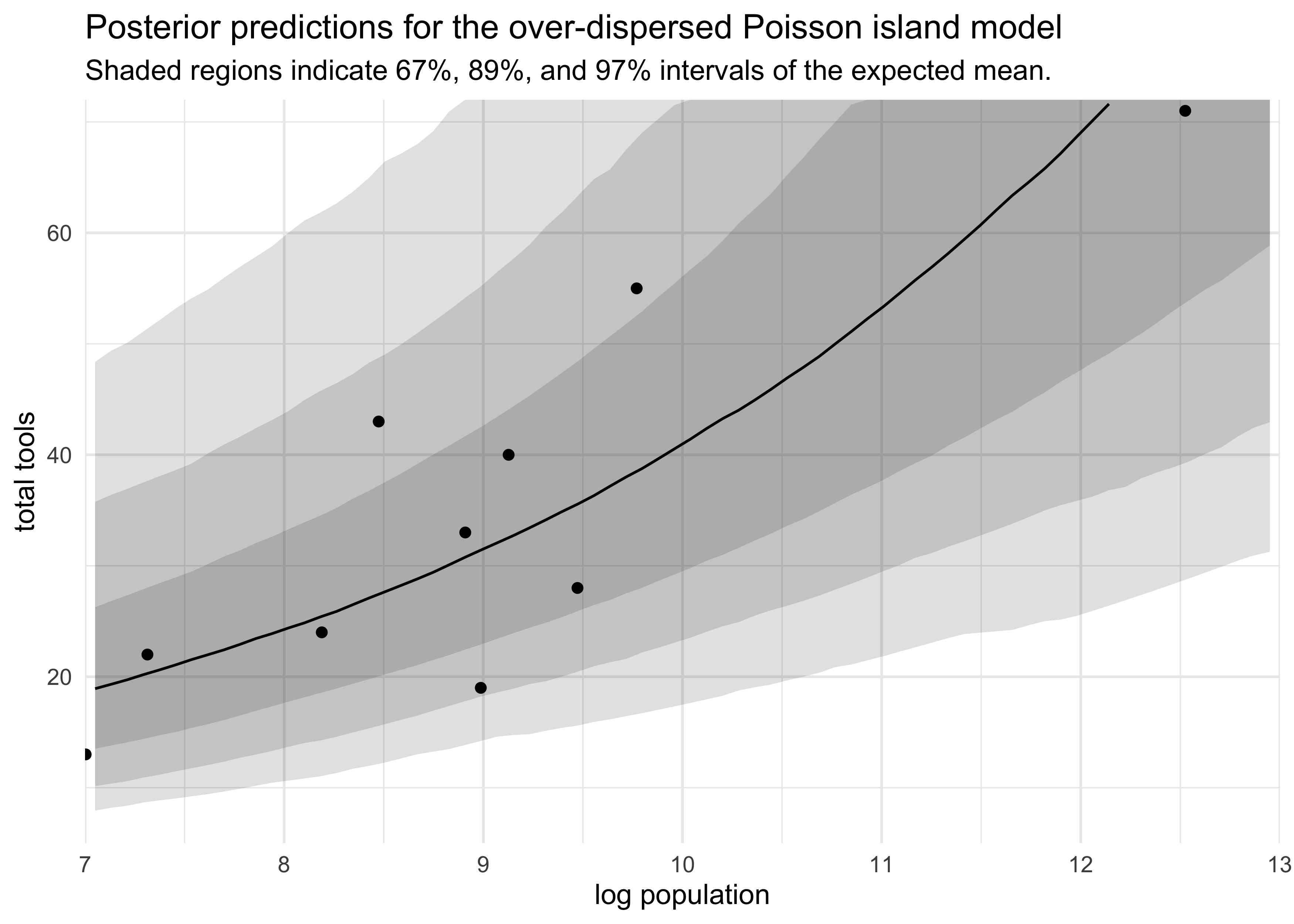
- plot posterior predictions that visualize the over-dispersion
- the
postcheck()function uses thea_societyvalues directly, not the hyperparametersaandsigma_societythat describe the dispersion - instead need to simulate counterfactual societies using these hyperparameters $\alpha$ and $\sigma_\text{society}$
- the
post <- extract.samples(m12_6)
d_pred <- tibble(
logpop = seq(6, 14, length.out = 100),
society = rep(1, 100)
)
# Sample possible alpha society values.
a_society_sims <- rnorm(2e4, mean = 0, post$sigma_society)
a_society_sims <- matrix(a_society_sims, nrow = 2e3, ncol = 10)
# Make predictions using the simulated a_society values.
link_m12_6 <- link(m12_6, n = 2e3, data = d_pred,
replace = list(a_society = a_society_sims))
#> [ 200 / 2000 ][ 400 / 2000 ][ 600 / 2000 ][ 800 / 2000 ][ 1000 / 2000 ][ 1200 / 2000 ][ 1400 / 2000 ][ 1600 / 2000 ][ 1800 / 2000 ][ 2000 / 2000 ]
d_pred_res <- d_pred %>%
mutate(mu_median = apply(link_m12_6, 2, median)) %>%
bind_cols(
apply(link_m12_6, 2, PI, prob = 0.67) %>% pi_to_df(),
apply(link_m12_6, 2, PI, prob = 0.89) %>% pi_to_df(),
apply(link_m12_6, 2, PI, prob = 0.97) %>% pi_to_df()
)
d_pred_res %>%
mutate(x84_percent = scales::squish(x84_percent, range = c(0, 72)),
x94_percent = scales::squish(x94_percent, range = c(0, 72)),
x98_percent = scales::squish(x98_percent, range = c(0, 72))) %>%
ggplot(aes(x = logpop)) +
geom_ribbon(aes(ymin = x2_percent, ymax = x98_percent),
alpha = 0.15) +
geom_ribbon(aes(ymin = x5_percent, ymax = x94_percent),
alpha = 0.15) +
geom_ribbon(aes(ymin = x16_percent, ymax = x84_percent),
alpha = 0.15) +
geom_point(aes(y = total_tools),
data = d) +
geom_line(aes(y = mu_median)) +
scale_x_continuous(limits = c(7, 13), expand = c(0, 0)) +
scale_y_continuous(limits = c(5, 72), expand = c(0, 0)) +
labs(x = "log population",
y = "total tools",
title = "Posterior predictions for the over-dispersed Poisson island model",
subtitle = "Shaded regions indicate 67%, 89%, and 97% intervals of the expected mean.")
#> Warning: Removed 36 row(s) containing missing values (geom_path).
12.6 Practice
Easy
12E2. Make the following model into a multilevel model.
$$ y_i \sim \text{Binomial}(1, p_i) $$ $$ \text{logit}(p_i) = \alpha + \alpha_{\text{group}[i]} + \beta x_i $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \beta \sim \text{Normal}(0, 1) $$ $$ \alpha_\text{group} \sim \text{Normal}(0, \sigma_\text{group}) $$ $$ \sigma_\text{group} \sim \text{HalfCauchy(0, 1)} $$