Chapter 11. Monsters And Mixtures
- build more types of models by piecing together types we have already
learned about
- will discuss ordered categorical models and zero-inflated/zero-augmented models
- mixtures are powerful, but interpretation is difficult
11.1 Ordered categorical outcomes
- the outcome is discrete and has different levels along a dimension
but the differences between each level are not necessarily equal
- is a multinomial prediction problem with a constraint on the order of the categories
- want an estimate of the effect of a change in a predictor on the change along the categories
- use a cumulative link function
- the cumulative probability of a value is the probability of that value or any smaller value
- this guarantees the ordering of the outcomes
11.1.1 Example: Moral intuition
- example data come from a survey of people with different versions of
the classic “Trolley problem”
- 3 versions that invoke different moral principles: “action principle,” “intention principle,” and “contact principle”
- the goal is how people just the different choices from the different principles
response: from an integer 1-7, how morally permissible is the action
data("Trolley")
d <- as_tibble(Trolley) %>%
rename(q_case = case)
11.1.2 Describing an ordered distribution with intercepts
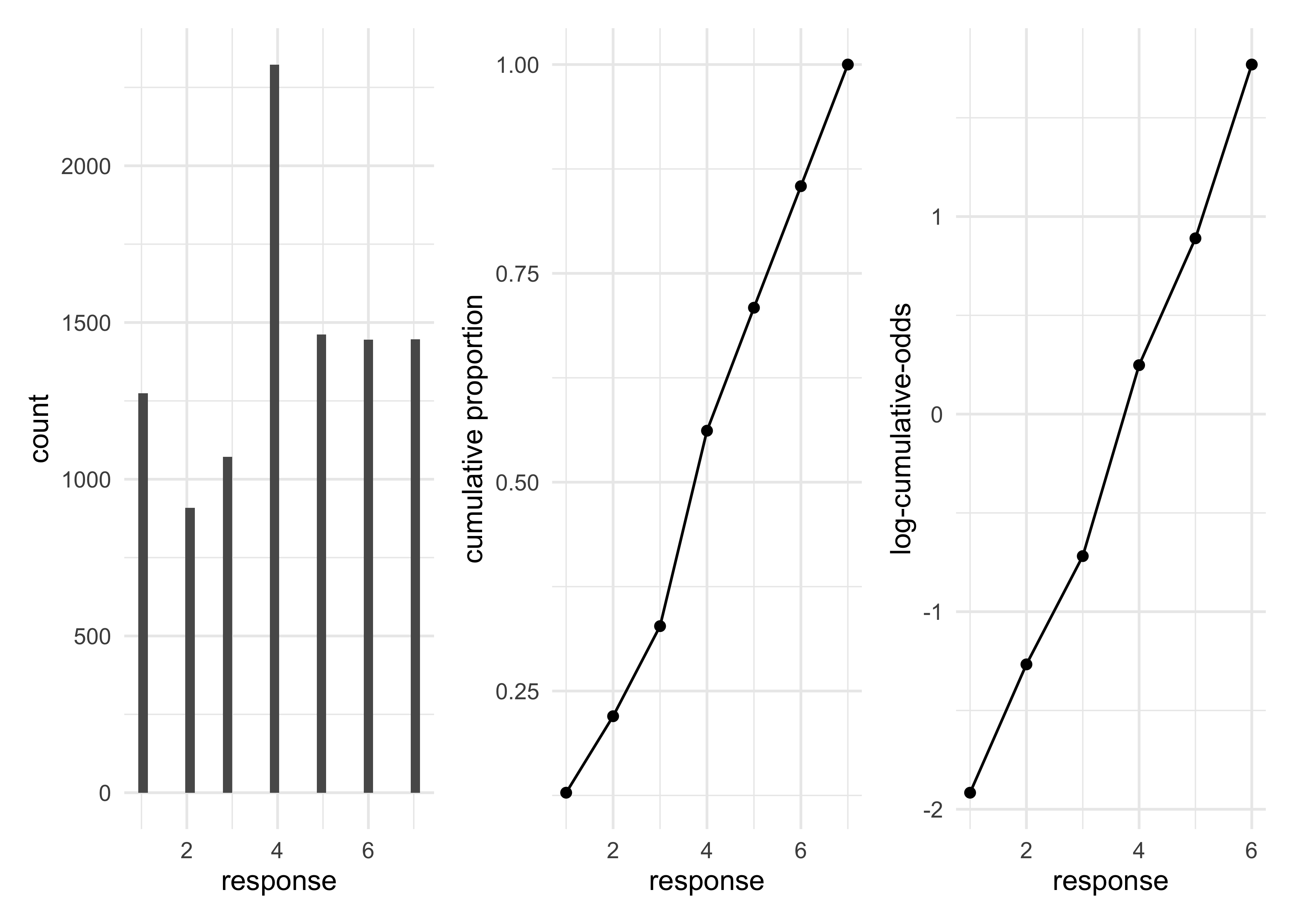
- some plots of the data
- a histogram of the response values
- cumulative proportion of responses
- log-cumulative-odds of responses
p1 <- d %>%
ggplot(aes(x = response)) +
geom_histogram(bins = 30) +
labs("Distribution of response")
p2 <- d %>%
count(response) %>%
mutate(prop = n / sum(n),
cum_prop = cumsum(prop)) %>%
ggplot(aes(x = response, y = cum_prop)) +
geom_line() +
geom_point() +
labs(y = "cumulative proportion")
p3 <- d %>%
count(response) %>%
mutate(prop = n / sum(n),
cum_prop = cumsum(prop),
cum_odds = cum_prop / (1 - cum_prop),
log_cum_odds= log(cum_odds)) %>%
filter(is.finite(log_cum_odds)) %>%
ggplot(aes(x = response, y = log_cum_odds)) +
geom_line() +
geom_point() +
labs(y = "log-cumulative-odds")
p1 | p2 | p3
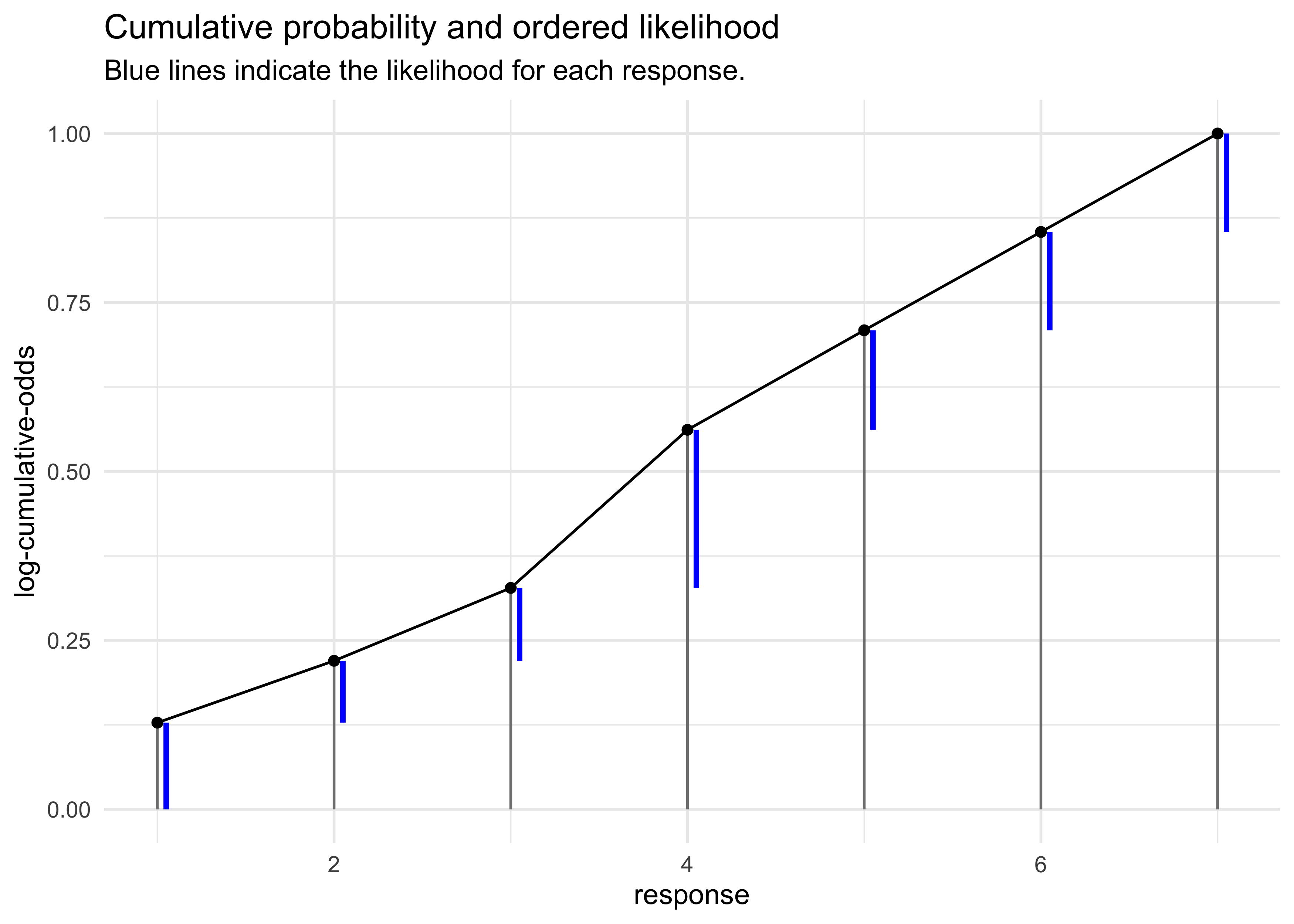
- why use the log-cumulative-odds of each response:
- it is the cumulative analog of the logit link used previously
- the logit is the log-odds; the cumulative logit is the log-cumulative-odds
- constrains the probabilities to between 0 and 1
- this link function takes care of converting the parameter estimates to the probability scale
- to use Bayes’ theorom to compute the posterior distribution of these
intercepts, need to compute the likelihood of each possible response
value
- need to use the cumulative probabilities $\Pr(y_i \ge k)$ to compute the likelihood $\Pr(y_i = k)$
- use the inverse link to translate the log-cumulative-odds back to cumulative probability
- therefore, when we observe $k$ and need its likelihoood, just
use subtraction:
- the values are shown as blue lines in the next plot
$$ p_k = \Pr(y_i = k) = \Pr(y_i \le k) - \Pr(y_i \le k - 1) $$
offset_subtraction <- function(x) {
y <- x
for (i in seq(1, length(x))) {
if (i == 1) {
y[[i]] <- x[[i]]
} else {
y[[i]] <- x[[i]] - x[[i - 1]]
}
}
return(y)
}
d %>%
count(response) %>%
mutate(prop = n / sum(n),
cum_prop = cumsum(prop),
likelihood = offset_subtraction(cum_prop),
ymin = cum_prop - likelihood) %>%
ggplot(aes(x = response)) +
geom_linerange(aes(ymin = 0, ymax = cum_prop), color = "grey50") +
geom_line(aes(y = cum_prop)) +
geom_point(aes(y = cum_prop)) +
geom_linerange(aes(ymin = ymin, ymax = cum_prop), color = "blue",
position = position_nudge(x = 0.05), size = 1) +
labs(y = "log-cumulative-odds",
title = "Cumulative probability and ordered likelihood",
subtitle = "Blue lines indicate the likelihood for each response.")
- below is the matematical form of the model using an ordered logit
likelihood
- notation for these models can vary by author
- the Ordered distribution is just a categorical distribution
that takes a vector
$\text{p} = {p_1, p_2, p_3, p_4, p_5, p_6}$
- only 6 because the 7th level has the value 1 automatically
- each response value $k$ gets an intercept parameter $\alpha_k$
$$ R_i \sim \text{Ordered}(p) $$ $$ logit(P_k) = \alpha_k $$ $$ \alpha_k \sim \text{Normal}(0, 10) $$
- the first model does not include any predictor vairables
- the link function is embedded in the likelihood function,
already
- simpler to type and makes the calculations more efficient, too
phiis a placeholder for now but will be used to add in predictor variables- the start values are included to start the intercepts in the
right order
- their exact values don’t really matter, just the order
- the link function is embedded in the likelihood function,
already
m11_1 <- quap(
alist(
response ~ dordlogit(phi, c(a1, a2, a3, a4, a5, a6)),
phi <- 0,
c(a1, a2, a3, a4, a5, a6) ~ dnorm(0, 10)
),
data = d,
start = list(a1 = -2, a2 = -1, a3 = 0, a4 = 1, a5 = 2, a6 = 2.5)
)
precis(m11_1)
#> mean sd 5.5% 94.5%
#> a1 -1.9160695 0.03000701 -1.9640265 -1.8681125
#> a2 -1.2666001 0.02423126 -1.3053263 -1.2278739
#> a3 -0.7186296 0.02137978 -0.7527986 -0.6844606
#> a4 0.2477844 0.02022442 0.2154619 0.2801070
#> a5 0.8898583 0.02208975 0.8545546 0.9251620
#> a6 1.7693642 0.02845011 1.7238954 1.8148329
- transform from log-cumulative-odds to cumulative probabilities
logistic(coef(m11_1))
#> a1 a2 a3 a4 a5 a6
#> 0.1283005 0.2198398 0.3276948 0.5616311 0.7088609 0.8543786
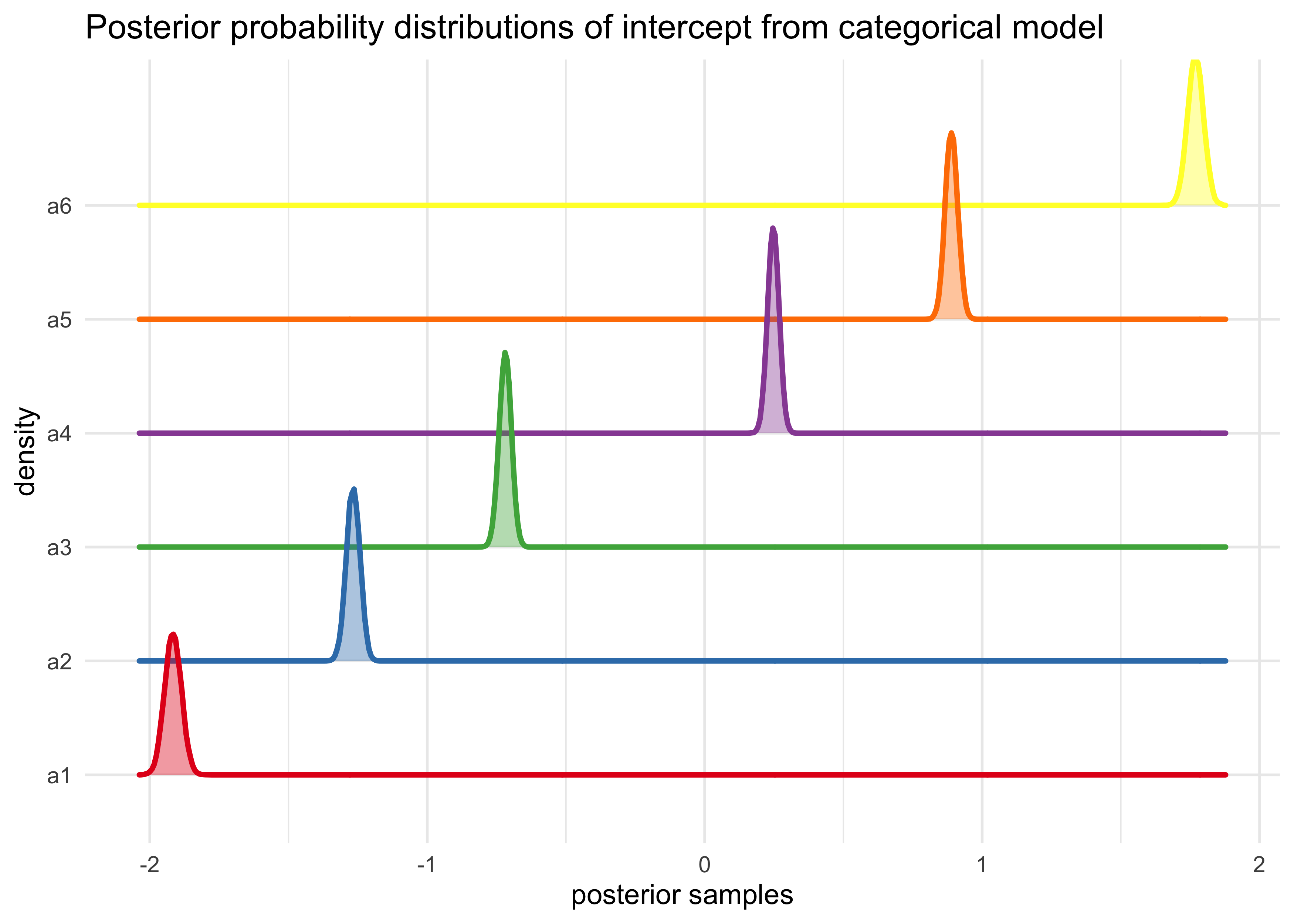
m11_1_link <- extract.samples(m11_1)
m11_1_link %>%
as.data.frame() %>%
as_tibble() %>%
pivot_longer(tidyselect::everything()) %>%
ggplot(aes(x = value, y = name, color = name, fill = name)) +
ggridges::geom_density_ridges(size = 1, alpha = 0.4, ) +
scale_color_brewer(palette = "Set1") +
scale_fill_brewer(palette = "Set1") +
theme(legend.position = "none") +
labs(x = "posterior samples",
y = "density",
title = "Posterior probability distributions of intercept from categorical model")
#> Picking joint bandwidth of 0.00348
11.1.3 Adding predictor variables
- to include predictor variables:
- define the log-cumulative-odds of each response $k$ as a sum of its intercept $\alpha_k$ and a typical linear model
- for example: add a predictor $x$ to the model
- define the linear model $\phi_i = \beta x_i$
- the cumulative logit becomes:
$$ \log \frac{\Pr(y_i \ge k)}{1 - \Pr(y_i \ge k)} = \alpha - \phi_i = \alpha - \beta x_i $$
- this form keeps the correct ordering of the outcome values while still morphing the likelihood of each individual value as the predictor $x_i$ changes value
- the linear model $\phi$ is subtracted from the intercept:
- because decreasing the log-cumulative-odds of every outcome value $k$ below the maximum shifts probability mass upwards towards higher outcome values
- a positive $\beta$ value indicates than an increase in the predictor variable $x$ results in an increase in the average response
- for the Trolly data, we can icnlude predictor variables for the
different types of questions: “action,” “intention,” and “contact”
- the formulation of the log-cumulative-odds of each response $k$ is shown below
- defines the log-odds of each possible response to be an additive model of the features of the story corresponding to each response
$$ \log \frac{\Pr(y_i \ge k)}{1 - \Pr(y_i \ge k)} = \alpha - \phi_i $$ $$ \phi_i = \beta_A A_i + \beta_I I_i + \beta_C C_i $$
m11_2 <- quap(
alist(
response ~ dordlogit(phi, c(a1, a2, a3, a4, a5, a6)),
phi <- bA*action + bI*intention + bC*contact,
c(a1, a2, a3, a4, a5, a6) ~ dnorm(0, 10),
c(bA, bI, bC) ~ dnorm(0, 10)
),
data = d,
start = list(a1 = -2, a2 = -1, a3 = 0, a4 = 1, a5 = 2, a6 = 2.5)
)
- fit another model with interactions between action and intention and
between contact and intention
- these two make sense in terms of the scenario we are modeling
while an interaction between contact and action does not
- contact is a type of action
- these two make sense in terms of the scenario we are modeling
while an interaction between contact and action does not
m11_3 <- quap(
alist(
response ~ dordlogit(phi, c(a1, a2, a3, a4, a5, a6)),
phi <- bA*action + bI*intention + bC*contact + bAI*action*intention + bCI*contact*intention,
c(a1, a2, a3, a4, a5, a6) ~ dnorm(0, 10),
c(bA, bI, bC, bAI, bCI) ~ dnorm(0, 10)
),
data = d,
start = list(a1 = -2, a2 = -1, a3 = 0, a4 = 1, a5 = 2, a6 = 2.5)
)
coeftab(m11_1, m11_2, m11_3)
#> m11_1 m11_2 m11_3
#> a1 -1.92 -2.84 -2.63
#> a2 -1.27 -2.15 -1.94
#> a3 -0.72 -1.57 -1.34
#> a4 0.25 -0.55 -0.31
#> a5 0.89 0.12 0.36
#> a6 1.77 1.02 1.27
#> bA NA -0.71 -0.47
#> bI NA -0.72 -0.28
#> bC NA -0.96 -0.33
#> bAI NA NA -0.45
#> bCI NA NA -1.27
#> nobs 9930 9930 9930
- interpretation:
- the intercepts are difficult to interpret on their own, but act
like regular intercepts in simpler models
- they are the relative frequencies of the outcomes when all predictors are set to 0
- there are 5 slope parameters: 3 main effects and 2 iinteractions
- they are all far from 0 (can check with
precis(m)) - they are all negative: each factor/interaction reduces the average response
- these values are difficult to interpret as is, so they are investigated more below
- they are all far from 0 (can check with
- the intercepts are difficult to interpret on their own, but act
like regular intercepts in simpler models
- compare the models by WAIC
- the model with interaction terms is sufficiently better than the other two, so we can safely proceed with just analyzing it and ignoring the other two
compare(m11_1, m11_2, m11_3)
#> WAIC SE dWAIC dSE pWAIC weight
#> m11_3 36929.15 81.16718 0.0000 NA 11.004379 1.000000e+00
#> m11_2 37090.36 76.34564 161.2159 25.78738 9.253143 9.826897e-36
#> m11_1 37854.49 57.63045 925.3444 62.65109 6.020406 1.158826e-201
- plot implied predictions to understand what model
m11_3implies- difficult to plot the predictions of log-cumulative-odds because each prediction is a vector of probabilities, one for each possible outcome
- as a predictor variable changes value, the entire vector changes
- one common plot is to use the horiztonal axis for the predictor
variable and the vertical axis for the cumulative probability
- plot a curve for each response value
post <- extract.samples(m11_3)
get_m11_3_predictions <- function(kA, kC, kI) {
res <- tibble()
for (s in 1:100) {
p <- post[s, ]
ak <- as.numeric(p[1:6])
phi <- p$bA * kA + p$bI*kI + p$bC*kC + p$bAI*kA*kI + p$bCI*kC*kI
pk <- pordlogit(1:6, a = ak, phi = phi)
res <- bind_rows(
res,
tibble(lvl = 1:6, val = pk)
)
}
return(res)
}
implied_predictions <- expand.grid(kA = 0:1, kC = 0:1, kI = 0:1) %>%
as_tibble() %>%
filter(!(kA == 1 & kC == 1)) %>%
group_by(kA, kC, kI) %>%
mutate(preds = list(get_m11_3_predictions(kA, kC, kI))) %>%
unnest(preds) %>%
ungroup()
implied_predictions %>%
mutate(facet = paste0("action: ", kA, ", ", "contact: ", kC)) %>%
group_by(kA, kC, kI) %>%
mutate(row_idx = row_number()) %>%
ungroup() %>%
mutate(line_group = paste(lvl, row_idx)) %>%
ggplot(aes(x = kI, y = val, group = line_group)) +
facet_wrap(~ facet, nrow = 1) +
geom_hline(yintercept = 0:1, lty = 2, color = "grey70", size = 1) +
geom_line(aes(color = factor(lvl)), alpha = 0.1) +
scale_color_brewer(palette = "Dark2") +
labs(x = "intention",
y = "probability",
color = "level",
title = "Implied predictions of model with interactions.")
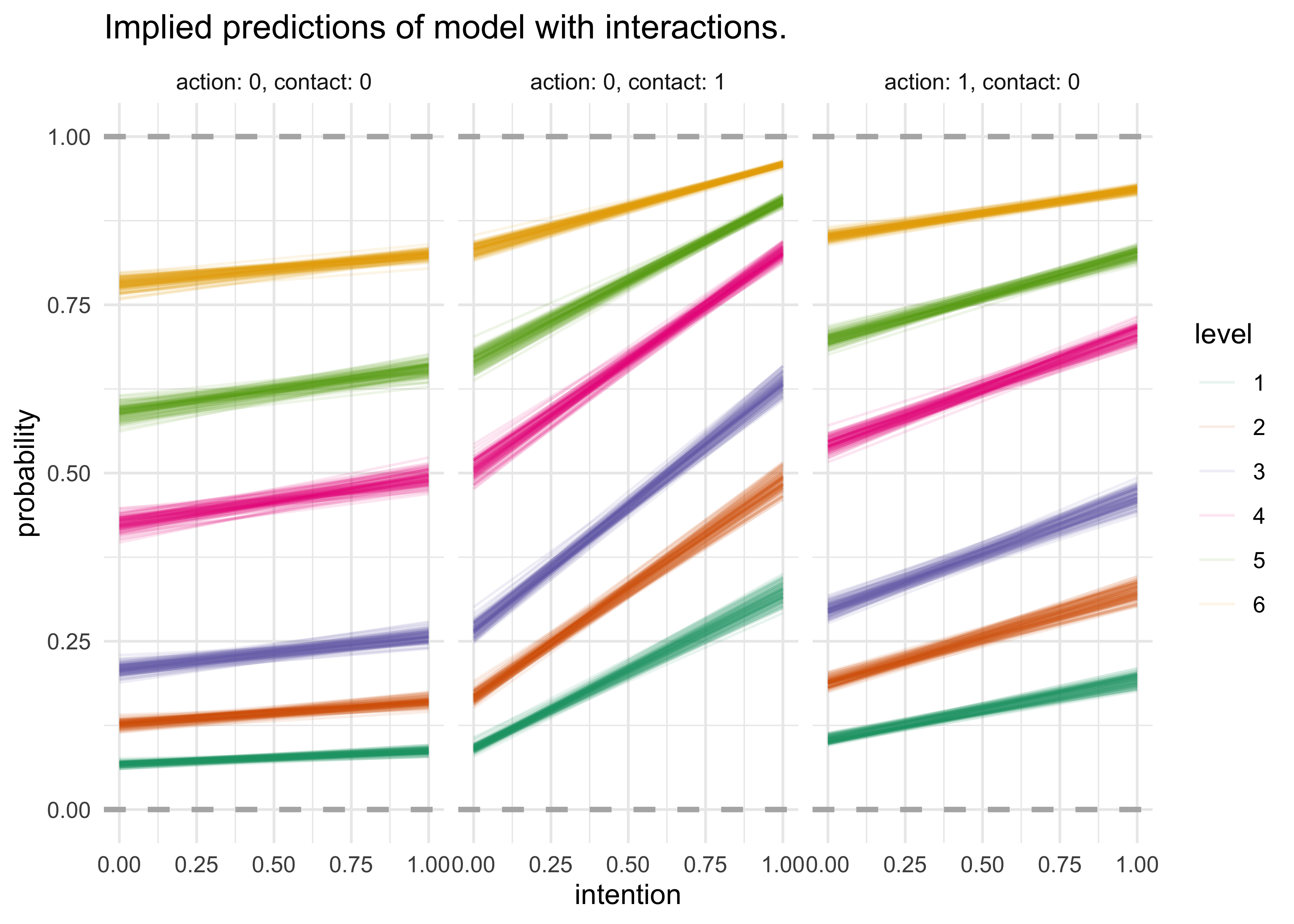
- interpretation:
- each horizontal line is the bounday between levels
- the thickness of the boundary represents the variation in prediction
- the change in height from intention changing from 0 to 1
indicates the predicted impact of changing a trolley story from
non-intention to intention
- the left-hand plot shows that there is not much change from switching intention when action and contact are both 0
- the other two plots show the interaction between intention and the other variable that is set to 1
- the middle plot shows that there is a large interaction between contact and intention
11.2 Zero-inflated outcomes
- mixture model: measurements arise from multiple proceesses;
different causes for the same observation
- uses more than one probability distribution
- common to need to use a mixture model for count variables
- a count of 0 can often arise in more than one way
- a 0 could occurr because nothing happens, because the rate of the event is very low, or because the event-generating process never began
- said to be zero-inflated
11.2.1 Example: Zero-inflated Poisson
- previously, we use the monastery example to study the Poisson
distribution
- now imagine that the monks take breaks on some days and no manuscripts are made
- want to estimate how often breaks are taken
- mixture model:
- a zero can arise from two processes:
- the monks took a break
- the monks workked but did not complete a manuscript
- let $p$ be the probability the monks took a break on a day
- let $\lambda$ be the mean number of manuscripts completed when the monks work
- a zero can arise from two processes:
- need a likelihood function that mixes these two processes:
- the following equation says this: “The probability of observing a zero is the probability that the monks took a break OR ($+$) the probability the monks worked AND ($\times$) failed to finish.”
$$ \Pr(0 | p, \lambda) = \Pr(\text{break} | p) + \Pr(\text{work} | p) \times \Pr(0 | \lambda) $$ $$ \Pr(0 | p, \lambda) = p + (1-p) e^{-\lambda} $$
- the likelihood of a non-zero value $y$ is below
- the the probability that the monks work multiplied by the probability that the working monks produce a manuscript
$$ \Pr(y | p, \lambda) = \Pr(\text{break} | p)(0) + \Pr(\text{work} | p) \Pr(y | \lambda) $$ $$ \Pr(y | p, \lambda) = (1 - p) \frac{\lambda^y e^{-\lambda}}{y!} $$
- can define
ZIPoissonas the distribution above with parameters $p$ (probability of 0) and $\lambda$ (mean of Poisson) to describe the shape- two linear models and two link functions, one for each process
$$ y_i \sim \text{ZIPoisson}(p_i, \lambda_i) $$ $$ \text{logit}(p_i) = \alpha_p + \beta_p x_i $$ $$ \log(\lambda_i) = \alpha_\lambda + \beta_\lambda x_i $$
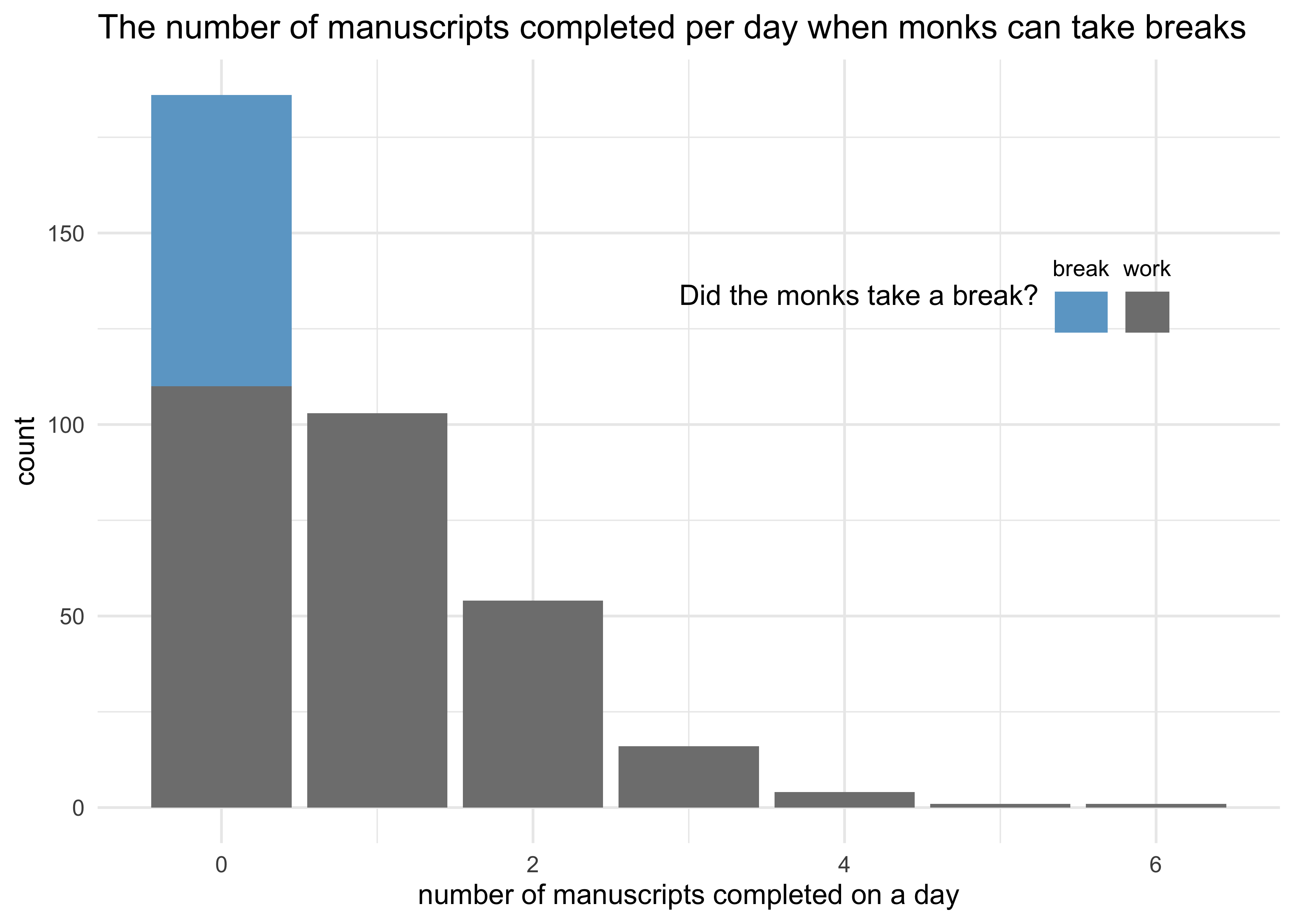
- need to simulate data for the monks taking breaks
# They take a break on 20% of the days
prob_break <- 0.2
# Average of 1 manuscript per working day.
rate_work <- 1
# Sample one year of production.
N <- 365
# Simulate which days the monks take breaks.
break_days <- rbinom(N, 1, prob = prob_break)
# Simulate the manuscripts completed.
y <- (1-break_days) * rpois(N, rate_work)
tibble(y, break_days) %>%
count(y, break_days) %>%
mutate(break_days = ifelse(break_days == 0, "work", "break"),
break_days = factor(break_days, levels = c("break", "work"))) %>%
ggplot(aes(x = y, y = n)) +
geom_col(aes(fill = break_days)) +
scale_fill_manual(
values = c("skyblue3", "grey50"),
guide = guide_legend(title.position = "left",
title.hjust = 0.5,
label.position = "top",
ncol = 2)
) +
theme(
legend.position = c(0.7, 0.7)
) +
labs(x = "number of manuscripts completed on a day",
y = "count",
fill = "Did the monks take a break?",
title = "The number of manuscripts completed per day when monks can take breaks")
- now we can fit a model
m11_4 <- quap(
alist(
y ~ dzipois(p, lambda),
logit(p) <- ap,
log(lambda) <- al,
ap ~ dnorm(0, 1),
al ~ dnorm(0, 10)
),
data = tibble(y)
)
precis(m11_4)
#> mean sd 5.5% 94.5%
#> ap -1.08474003 0.27298308 -1.52101972 -0.6484603
#> al 0.04329227 0.08613731 -0.09437178 0.1809563
logistic(m11_4@coef[["ap"]])
#> [1] 0.2526101
exp(m11_4@coef[["al"]])
#> [1] 1.044243
- can get a very accurate prediction for the proportion of days taken
off by the monks and the rate of manuscript production per working
day
- though, cannot determine whether the monks took any particular day off
11.3 Over-dispersed outcomes
- over-dispersion: the variance of a variable exceededs the expected
amount for a model
- e.g.: for a binomial, the expected value is $np$ and its variance $np(1-p)$
- for a count model, this suggests that a necessary variable has been omitted
- ideally, would just include the missing variable to remove over-dispersion, but not always the case/possible
- two strategies for mitigating the effects of over-dispersion
- use a continuous mixture model: a linear model is attached to
a distribution of observations
- common models: beta-binomial and gamma-Poisson (negative-binomial)
- these are demonstrated in the following sections
- employ a multilevel model (GLMM) and estimate the residuals of
each observation and the distribution of those residuals
- easier to fit than beta-binmial and gamma-Poisson GLMs
- more flexible
- handle over-dispersion and other kinds of heterogineity simulatneously
- GLMMs are covered in the next chapter
- use a continuous mixture model: a linear model is attached to
a distribution of observations
11.3.1 Beta-binomial
- beta-binomial models assumes that each binomial count observation
has its own probability of a success
- estimates the distribution of probabilities of success across
cases
- instead of a single probability of success
- predictor variables change the shape of this distribution instead of directly determining the probability of each success
- estimates the distribution of probabilities of success across
cases
- example: UCB admissions data
- if we ignore the department, the data is very over-dispersed
- because the departments vary a lot in baseline admission rates
- therefore, ignoring the inter-department variation results in an incorrect inference about applicant gender
- can fit a beta-binomial model, ignoring department
- if we ignore the department, the data is very over-dispersed
- a beta-binomial model will assume that each row of the data has a
unique, unobserved probability of admission
- these probabilities of admission have a common distribution
described by the beta distribution
- use the beta distribution because it can be used to calculate the likelihood function that averages over the unknown probabilities for each observation
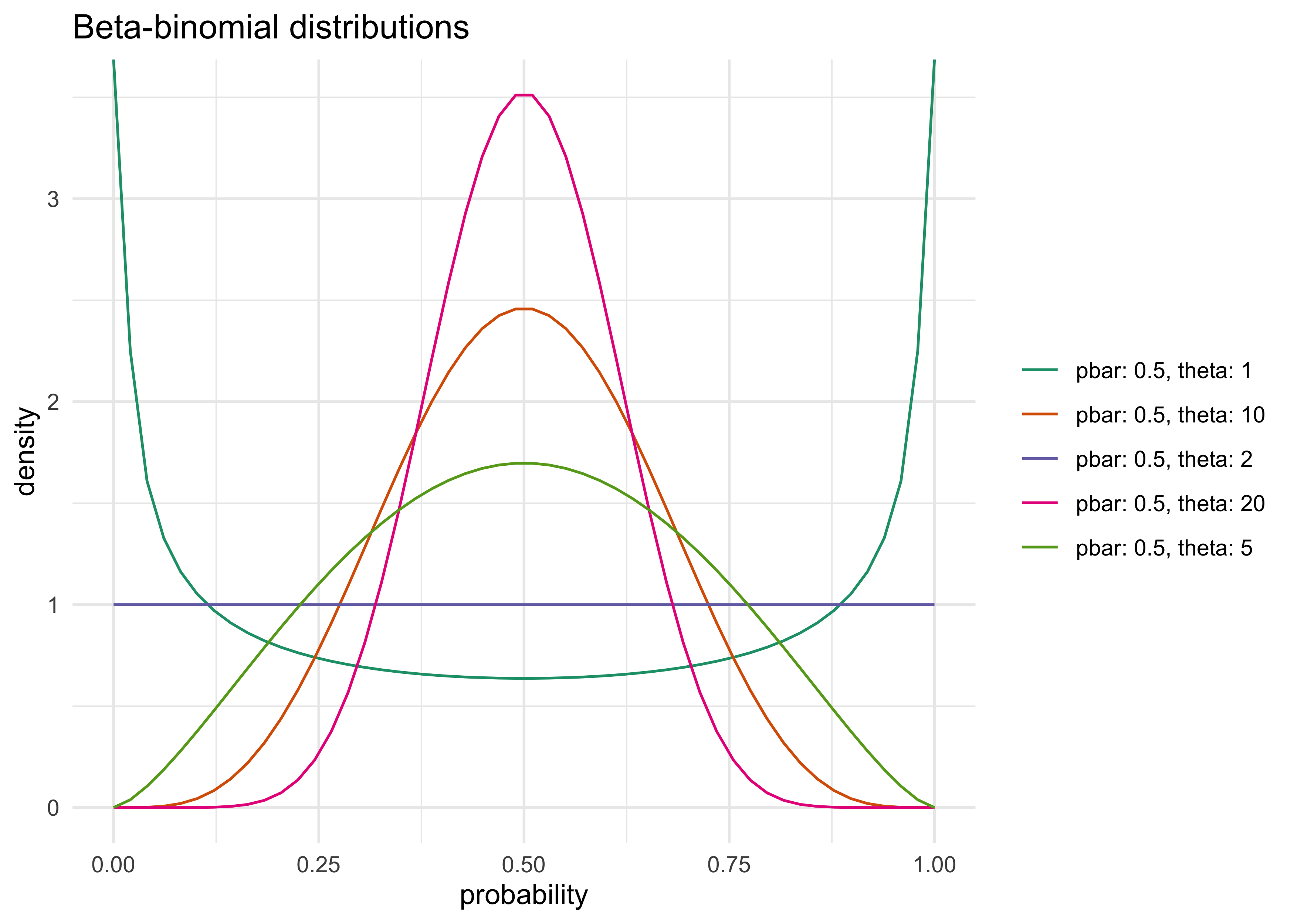
- beta-distribution has 2 parameters:
- $\bar{\textbf{p}}$: average probability
- $\theta$: shape parameter
- shape parameter describes the spread of the distribution
- these probabilities of admission have a common distribution
described by the beta distribution
pbar <- 0.5
thetas <- c(1, 2, 5, 10, 20)
x_vals <- seq(0, 1, length.out = 50)
beta_dist_res <- tibble()
for (theta in thetas) {
beta_dist_res <- bind_rows(
beta_dist_res,
tibble(theta = theta,
pbar = pbar,
x = x_vals,
d = dbeta2(x_vals, pbar, theta))
)
}
beta_dist_res %>%
mutate(params = paste0("pbar: ", pbar, ", theta: ", theta)) %>%
ggplot(aes(x = x, y = d)) +
geom_line(aes(group = params, color = params)) +
scale_color_brewer(palette = "Dark2") +
labs(x = "probability", y = "density", color = NULL,
title = "Beta-binomial distributions")
- we will build a linear model to $\bar{\textbf{p}}$ so that changes
in predictor variables change the central tendency of the
distribution
- $A$ is the number of admissions (
admitcolumn) - $n$ is the number of applications (
applicationscolumn) - any predictor variables could be included in the linear model for $\bar{p}$
- $A$ is the number of admissions (
$$ A_i \sim \text{BetaBinomial}(n_i, \bar{p}_i, \theta) $$ $$ \text{logit}(\bar{p}_i) = \alpha $$ $$ \alpha \sim \text{Normal}(0, 10) $$ $$ \theta \sim \text{HalfCauchy}(0, 1) $$
data("UCBadmit")
d <- as_tibble(UCBadmit) %>%
janitor::clean_names()
stash("m11_5", depends_on = "d", {
m11_5 <- map2stan(
alist(
admit ~ dbetabinom(applications, pbar, theta),
logit(pbar) <- a,
a ~ dnorm(0, 2),
theta ~ dexp(1)
),
data = d,
constraints = list(theta = "lower=0"),
start = list(theta = 3),
iter = 4e3,
warmup = 1e3,
chains = 2,
cores = 1
)
})
#> Loading stashed object.
plot(m11_5)
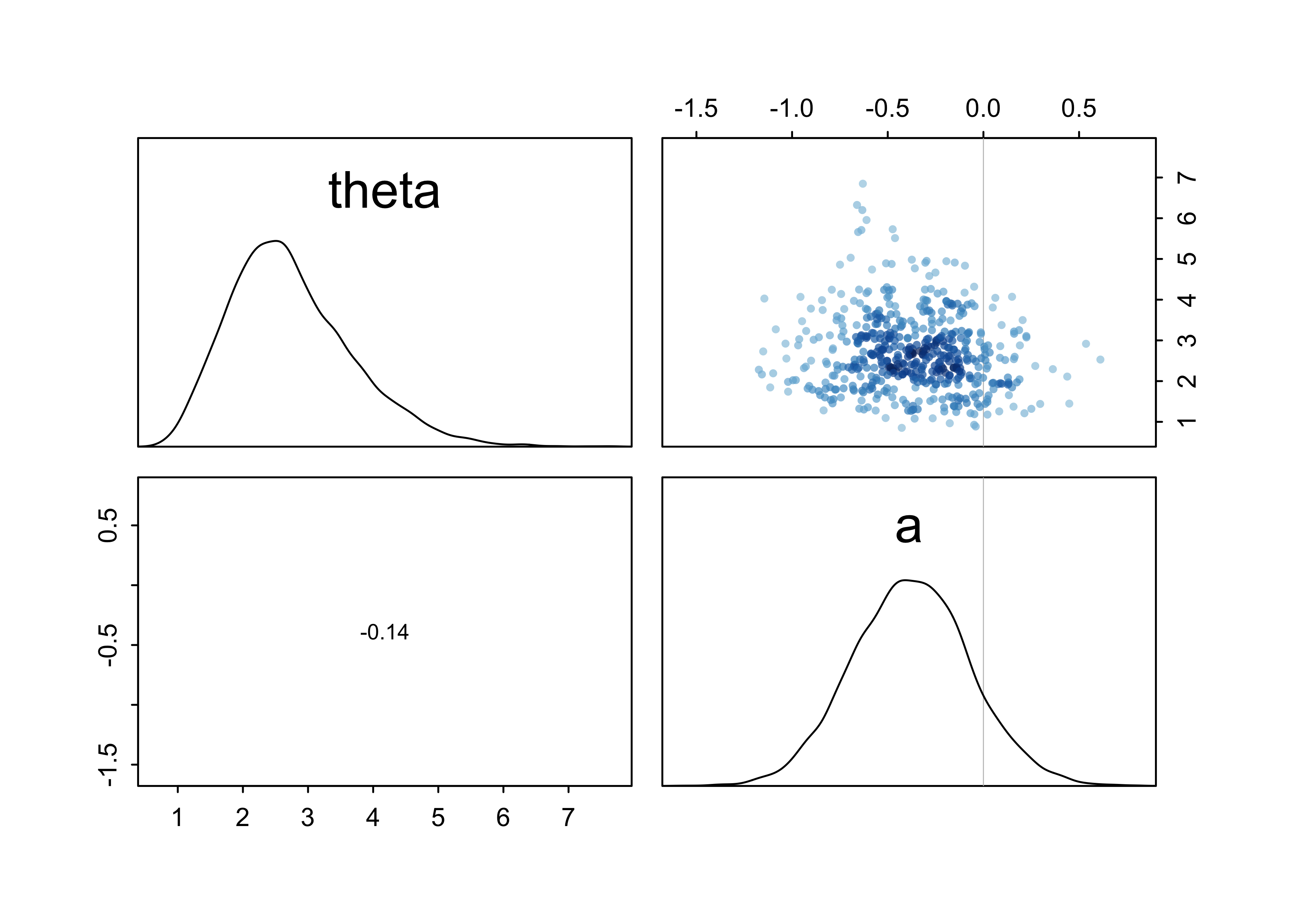
pairs(m11_5)
precis(m11_5)
#> mean sd 5.5% 94.5% n_eff Rhat4
#> theta 2.7399242 0.946932 1.4355572 4.4392494 3913.286 0.9997085
#> a -0.3775653 0.307988 -0.8717805 0.1203542 3450.286 1.0000152
- interpretation:
ais on the log-odds scale and defines $\bar{\textbf{p}}$ of the beta distribution of probabilities for each row of the data- therefore, the average probability of admission across departments is about 0.4, but the percentile is quite wide
post <- extract.samples(m11_5)
quantile(logistic(post$a), c(0.025, 0.5, 0.975))
#> 2.5% 50% 97.5%
#> 0.2749293 0.4066962 0.5588299
- to see what the model says of the data, need to account for
correlation between $\bar{\textbf{p}}$ and $\theta$
- these parameters define a distribution of distributions
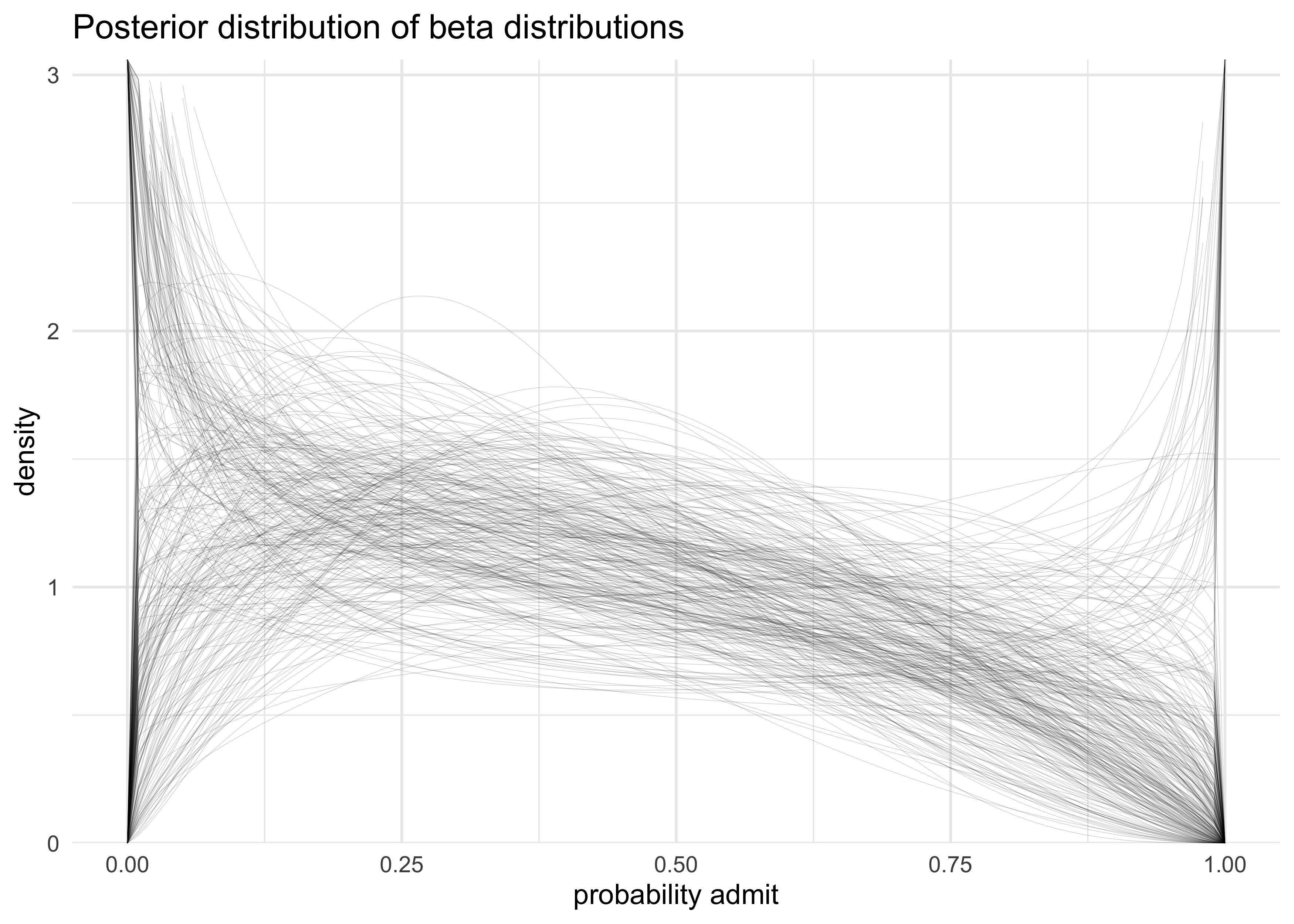
- 100 combinations of $\bar{\textbf{p}}$ and $\theta$ are shown in the following plot
dist_of_dist <- map(1:300, function(i) {
p <- logistic(post$a[i])
theta <- post$theta[i]
x <- seq(0, 1, length.out = 100)
probs <- dbeta2(x, p, theta)
return(tibble(i, p, theta, x, prob = probs))
}) %>%
bind_rows()
dist_of_dist %>%
ggplot(aes(x, prob)) +
geom_line(aes(group = factor(i)), size = 0.1, alpha = 0.2) +
scale_y_continuous(limits = c(0, 3),
expand = expansion(mult = c(0, 0.02))) +
labs(x = "probability admit",
y = "density",
title = "Posterior distribution of beta distributions")
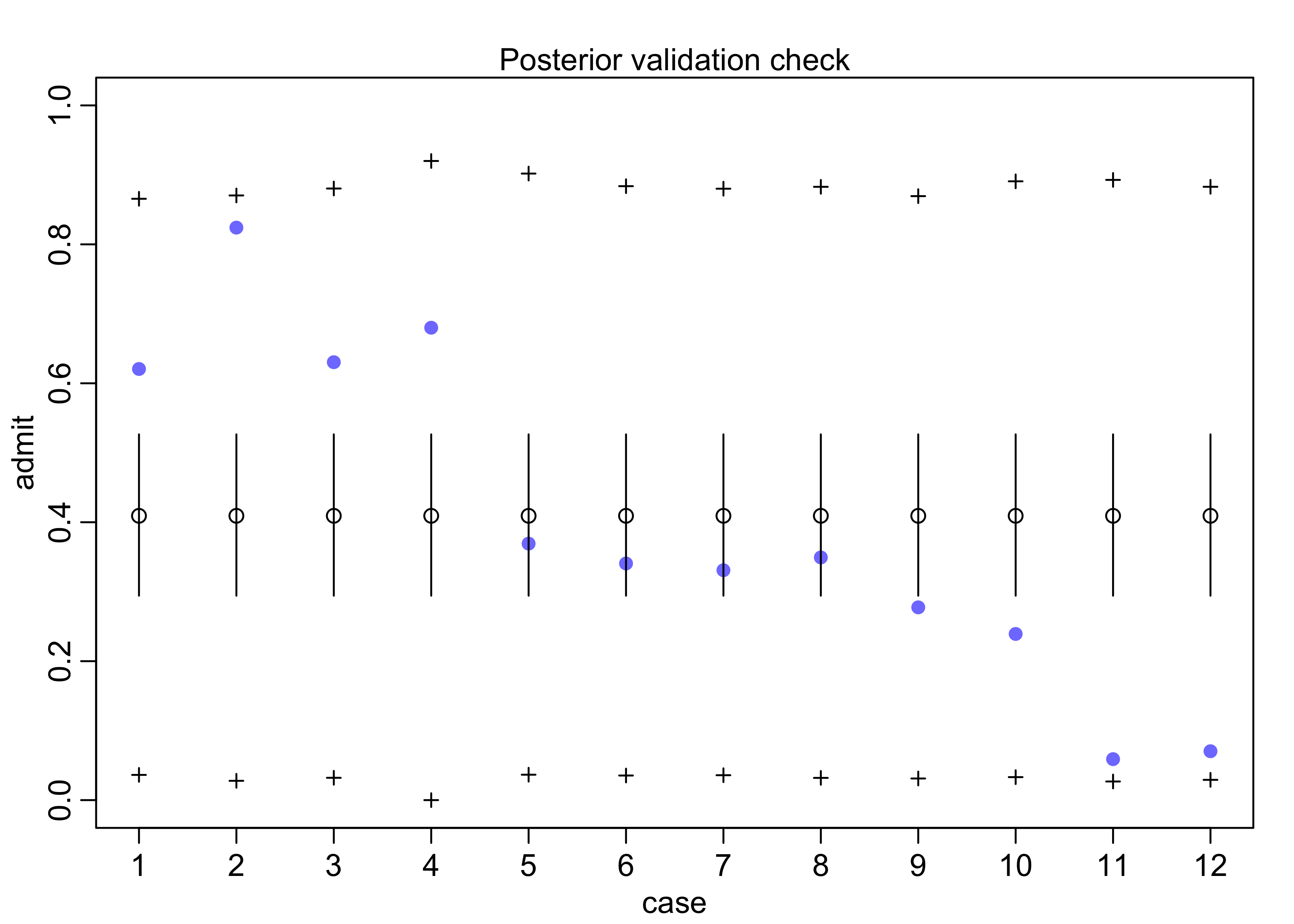
- use posterior check to see how the beta distribution of
probabilities of admissions influences predicted counts of
applications admitted
- the y-axis shows the predicted number of admits per row of the data frame on the x-axis
- the purple dots are the actual values
- open circles are the estimates from the model with 89% intervals
- the
+are the 89% interval of predicted counts of admission- shows there is a lot of dispersion
- this is from the different departments, but the model doesn’t know about them
- the beta distribution accounts for this heterogeneity
postcheck(m11_5)
#> [ 100 / 1000 ][ 200 / 1000 ][ 300 / 1000 ][ 400 / 1000 ][ 500 / 1000 ][ 600 / 1000 ][ 700 / 1000 ][ 800 / 1000 ][ 900 / 1000 ][ 1000 / 1000 ]
#> [ 100 / 1000 ][ 200 / 1000 ][ 300 / 1000 ][ 400 / 1000 ][ 500 / 1000 ][ 600 / 1000 ][ 700 / 1000 ][ 800 / 1000 ][ 900 / 1000 ][ 1000 / 1000 ]
11.3.2 Negative-binomial or gamma-Poisson
- negative-binomial (or gamma-Poisson): assumes that each Poisson
count observation has its own rate
- assumes the shape of the gamma distribution to describe the Poisson rates
- predictor variables adjust the shape of this distribution, not the expected value of each observation
- use the gamma distribution because it makes the math easier
- fitting a gamma-Poisson uses the
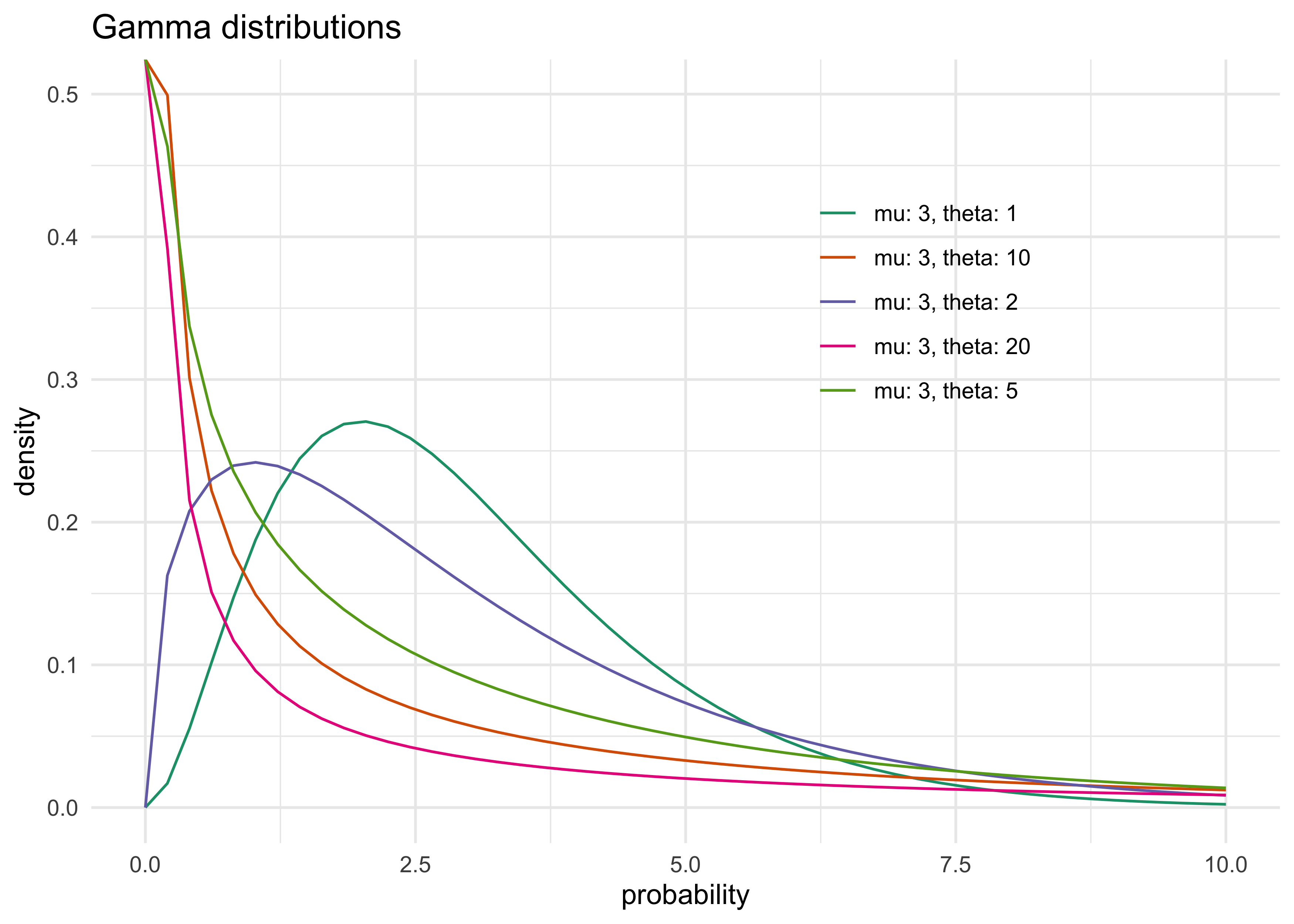
dgampois()function- distribution is defined by a mean $\mu$ and scale $\theta$
- as $\theta$ increases, the gamma distribution becomes for dispersed around the mean
- below are some examples of gamma distributions
- as $\theta$ approaches 0, the gamma approaches a Gaussian
mu <- 3
thetas <- c(1, 2, 5, 10, 20)
x_vals <- seq(0, 10, length.out = 50)
gamma_dist_res <- tibble()
for (theta in thetas) {
gamma_dist_res <- bind_rows(
gamma_dist_res,
tibble(theta,
mu,
x = x_vals,
d = dgamma2(x_vals, mu, theta))
)
}
gamma_dist_res %>%
mutate(params = paste0("mu: ", mu, ", theta: ", theta)) %>%
ggplot(aes(x = x, y = d)) +
geom_line(aes(group = params, color = params)) +
scale_color_brewer(palette = "Dark2") +
theme(
legend.position = c(0.7, 0.7)
) +
labs(x = "probability", y = "density", color = NULL,
title = "Gamma distributions")
- for fitting:
- a linear model can be attached to the definition of $\mu$ using the log link function
- there are examples in
?dgampoisand practice problems at the end of the chapter
11.3.3 Over-dispersion, entropy, and information criteria
- should use DIC instead of WAIC
- see text for an explination of why